Chapter: Genetics and Molecular Biology: Protein Synthesis
Termination, Nonsense, and Suppression - Protein Synthesis
Termination, Nonsense, and Suppression
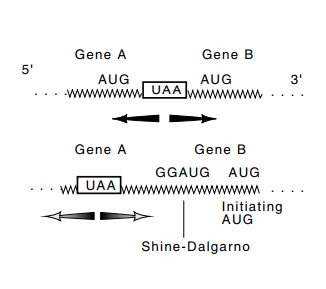
Figure
7.15 Translation termina-tion and
reinitiation. Top: A ribosome terminating at the UAA codon could directly
reinitiate at either of the nearby AUG codons if either possesses a ribosome
bind-ing site. Bottom: A ribosome terminating at the UAA codon would not be
able to drift to the more distant AUG codon on the right. A new initiation
complex would have to form using the GGAG sequence to bind 16S rRNA.
short distances and can reinitiate translation if
it encounters a ribo-some-binding site and an initiation codon before it
dissociates (Fig. 7.15).
Three proteins are involved in termination: a
protein factor R1, which is necessary for termination at UAA and UAG
codons; a protein factor R2, which is necessary for termination at UAA and UGA
codons; and a
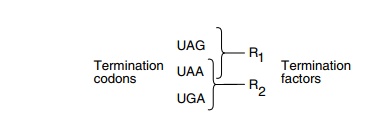
protein R3, which accelerates the
termination process. Cells contain approximately one molecule of R per five
ribosomes, a number consis-tent with the usage of these molecules.
The existence of chain termination codons is
responsible for an interesting phase in the growth of molecular biology. A
mutation within a gene can change one of the 61 sense codons into one of the
polypeptide chain terminating, or nonsense codons. This result shows that only
the three bases are necessary to code for chain termination, no others are
required, and no special secondary structure of the mRNA is required. As a
result of a nonsense codon within a gene, the protein encoded by the mutated
gene will be prematurely terminated during translation. Usually the shortened
polypeptide possesses no enzymatic activity, and it is frequently degraded by
proteases within the cell. An additional effect of a nonsense mutation is
depressed translation of a following gene in an operon. This polar effect
results from termination of tran-scription due to the sizeable portion of the
barren mRNA following the nonsense codon and preceding the next ribosome
initiation site. Quite surprisingly, some bacterial strains were found that
could suppress the effects of a nonsense mutation. Although the suppressors
rarely restored the levels of the “suppressed” protein to prior levels, the
cell often
possessed sufficient amounts of the protein to
survive. In the case of a nonsense mutation in a phage gene, the suppressor
strains permitted the phage to grow and form plaques.
Capecchi and Gussin showed that a suppressing
strain inserted a particular amino acid at the site of the nonsense mutation.
It did so by “mistranslating” the nonsense codon as a codon for an amino acid.
They also showed that the mistranslation resulted from a change in one of the
tRNAs for the inserted amino acid. Subsequently, sequencing of suppressor tRNAs
has shown that, except in a special case, their antico-dons have been altered
so as to become complementary to one of the termination codons. Apparently one
of two different events can then occur when a ribosome reaches a nonsense codon
in a suppressing strain. Termination can occur via the normal mechanism, or an
amino acid can be inserted into the growing polypeptide chain and translation
can proceed.
Using genetic selections, suppressors have been
found that insert tyrosine, tryptophan, leucine, glutamine, and serine. Except
in unusual cases, such suppressors must be derived from the original
tRNAs by single nucleotide changes. By chemical means and the utilization of
genetic engineering, an additional half dozen or so suppressors have been
synthesized.
The
termination codon UAG has come to be called amber and UAA called ochre. No
generally used name for the UGA codon exists, al-though it is sometimes called
opal. Amber-suppressing tRNAs read only the UAG codon, and ochre-suppressing
tRNAs read both UAA and UAG codons as a result of the “wobble” in translation.
Since the R factors are protein and cannot be constructed like tRNA, it is no
surprise that R2 does not “wobble” and does not recognize the UGG
(trp) codon.
How do
normal proteins terminate in suppressing cells? If a suppres-sor were always to
insert an amino acid in response to a termination codon instead of terminating,
then many of the cellular proteins in suppressor-containing cells would be
fused to other proteins or at least be appreciably longer than usual. The
problem of terminating normal proteins could be solved in part by the presence
of several different termination signals at the end of every gene. Then only
the introduction of several different suppressors to a cell could create
problems. Few genes, however, have been found to be ended by tandem translation
terminators.
The more
likely explanation for the viability of nonsense-suppressing strains is that
the efficiency of suppression never approaches 100%. Typically it is 10% to
40%. Hence suppression of normal translation termination codons can fuse or
lengthen some proteins in a cell, but most terminate as usual. On the other
hand, the gene possessing the nonsense mutation would occasionally yield a
suppressed instead of terminated protein. A suppression efficiency of 20% could
reduce the amount of some cellular proteins from 100% to 80%, relatively
speaking, not a substantial reduction. On the other hand, the existence of this
suppressor would raise the amount of the suppressed protein from 0% to 20% of normal. Relative to the nonsuppressed
level, this is an enormous increase.
Related Topics