Chapter: Genetics and Molecular Biology: Protein Synthesis
Protein Elongation Rates - Protein Synthesis
Protein Elongation Rates
In bacteria, the protein elongation rate is about
16 amino acids per second. This means the ribosomes are moving about 48
nucleotides per second along the messenger RNA. This value is very close to the
corresponding transcription rate of 50 to 60 bases per second. Therefore once a
ribosome begins translation it can keep up with the transcribing RNA
polymerase. In several of the better-studied operons, the rate of ribosome
attachment is sufficiently fast that the ribosomes are rather
How can the in
vivo rate of protein elongation be measured? One approach would be to use
methods analogous to those used to measure RNA elongation rates as described.
Unfortunately, no adequate inhibitors of protein initiation analogous to
rifamycin are known, and slightly more complicated experiments must be done.
The most general method for rate measurement uses an idea originally developed
for measurement of RNA elongation rates before rifamycin was available.
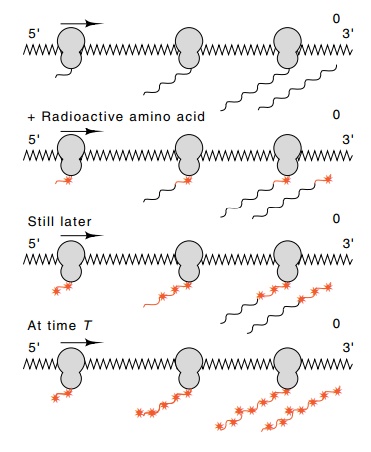
Figure 7.18 Synthesis of a pro-tein by ribosomes traversing a messenger. Ribosomes initiate at the 5’ end of the messenger, and the protein is completed and re-leased at the 3’ end. Addition of radioactive amino acids pro-duces partially labeled polypeptides, indicated by the stars, and a completed protein with radioactive amino acids near its carboxy terminus. Longer labeling intervals lead to longer radioactive regions until radioactive amino acids have been present for the length of time, T, required to synthesize the complete protein. After this time all released polypeptides are labeled over their entire lengths.
Consider an experiment in which a radioactive amino
acid is added to a culture of growing cells (Fig. 7.18). Let us focus our
attention on a class of protein of one particular size and consider intervals
that are short compared with the cell doubling time. The number of polypeptide
chains in the size class completed after the addition of radioactive label is
proportional to the time of labeling, nαt.
Also, for labeling intervals shorter than the time
required to synthe-size this size of polypeptide chain from one end to the
other, the average amount of label incorporated into each chain is proportional
to the time that label has been present. Hence during this early period, the
total amount of radioactivity in the particular size class increases in
propor-tion to the number of chains released multiplied by their average
radioactivity. Both of these are proportional to the time of labeling, t.
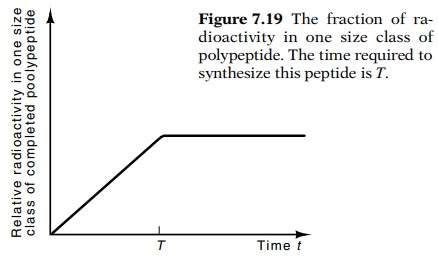
Figure
7.19 The fraction of ra-dioactivity in
one size class of polypeptide. The time required to synthesize this peptide is T.
Thus the radioactivity in the particular size class
of protein increases in proportion to t2.
After label has been present for the length of
time, T, necessary to synthesize
completely the polypeptide, the radioactivity per completed peptide chain can
no longer increase. After this time, the radioactivity in the particular
polypeptide size class can only increase in proportion to the number of chains
completed. That is, the radioactivity now increases in proportion to t.
For experimental quantitation, it is convenient to
compare the radio-activity in a particular size class of polypeptide to the
total radioactivity in all sizes of polypeptide. The total amount of
radioactivity in all size classes of polypeptide must increase in proportion to
the time of labeling, t. Therefore
the fraction of radioactivity in a particular size class of protein increases
in proportion to t until the
radioactive label has been present long enough for the entire length of the
protein to become radioactively labeled. Thereafter, the fraction of label in
the size class remains constant (Fig. 7.19). Hence, determining the transition
time from linear increase to being constant provides the synthesis time for the
particular size class of polypeptide.
The experimental protocol for performing the
elongation rate meas-urement is simply to add a radioactive amino acid to a
growing culture of cells. At intervals thereafter, samples are withdrawn and
the protein
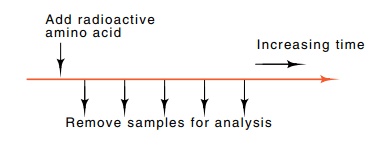
from the entire sample is denatured with sodium
dodecyl sulfate and electrophoresed on a polyacrylamide gel. This provides the
requisite size separation of the polypeptides as described.
Related Topics