Chapter: Genetics and Molecular Biology: Protein Synthesis
Decoding the Message - Protein Synthesis
Decoding the Message
What decodes the messages? Clearly, base pairing between a codon of message and an anticodon of the aminoacyl-tRNA decodes. This is not the complete story however. Since the ribosome pays no attention to the correctness of the tRNA charging, the aminoacyl-tRNA synthetases are just as important in decoding, for it is also essential that the tRNA molecules be charged with the correct amino acid in the first place.
Once the amino acids have been linked to their
cognate tRNAs, the process of protein synthesis shifts to the ribosome. The
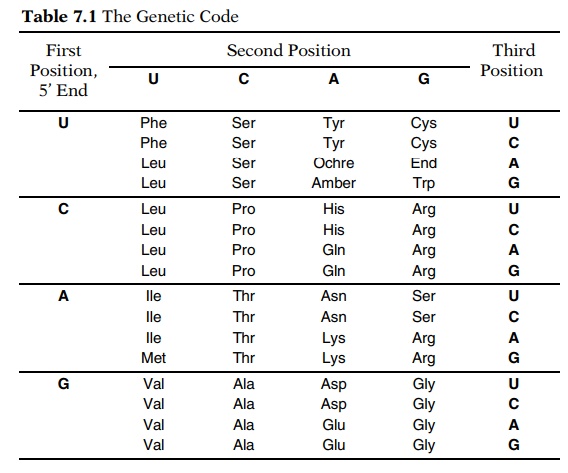
“code” is the correspondence between the triplets of bases in the codons and
the amino acids they specify (Table 7.1). In a few exciting years molecular
biologists progressed from knowing that there must be a code, to learning that
each amino acid is encoded by three bases on the messen-ger, to actually determining
the code. The history and experiments of the time are fascinating and can be
found in The Eighth Day of Creation
by Horace Freeland Judson.
In the later stages of solving the code it became
apparent that the code possessed certain degeneracies. Of the 64 possible
three-base codons, in most cells, 61 are used to specify the 20 amino acids.
One to six codons may specify a particular amino acid. As shown in Table 7.1,
synonyms generally differ in the third base of the codons. In the third
position, U is equivalent to C and, except for methionine and trypto-phan, G is
equivalent to A.
Under special circumstances likely to reflect early
evolutionary his-tory, one of the three codons that code for polypeptide chain
termina-tion also codes for the insertion of selenocysteine. The same codon at
the end of other genes specifies chain termination. Thus, the context
surrounding this codon also determines how it is read. Generally such
With the study of purified tRNAs, several facts
became apparent. First, tRNA contains a number of unusual bases, one of which
is inosine,
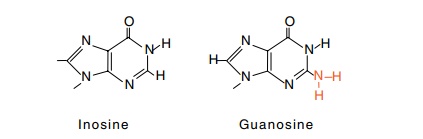
which is occasionally found in the first position
of the anticodon. Second, more than one species of tRNA exists for most of the
amino acids. Remarkably, however, the different species of tRNA for any amino
acid all appear to be charged by the same synthetase. Third, strict
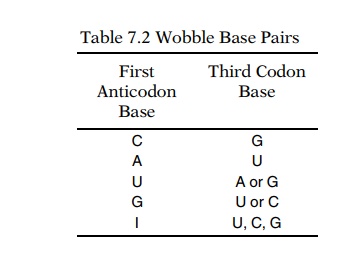
Watson-Crick base pairing is not always followed in the third position of the
codon (Table 7.2). Apparently the third base pair of the codon-anticodon
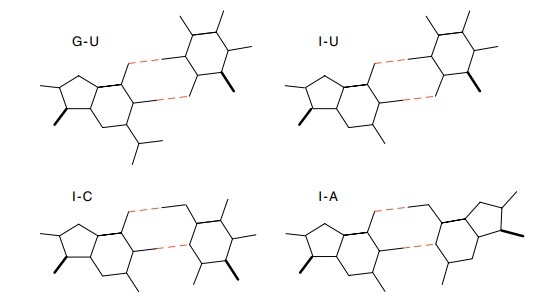
complex permits a variety of base pairings (Fig. 7.7). This phenomenon is
called wobble. The G-U base pair of alanine tRNA mentioned above is an example
of wobble.
Figure
7.7 Structure of the wobble base
pairs.
In mitochondria the genetic code might be slightly
different from that described above. There the translation machinery appears
capable of translating all the codons used with only 22 different species of
tRNA. One obstacle to understanding translation in mitochondria is the RNA
editing that can occur in these organelles. Because the sequence of mRNA can be
changed after its synthesis, we cannot be sure that the sequence of genes as
deduced from the DNA is the sequence that actually is translated at the
ribosome. Hence deductions about the use or lack of use of particular codons as
read from the DNA sequence cannot be made with reliability.
Related Topics