Chapter: Genetics and Molecular Biology: Protein Synthesis
Eukaryotic Translation and the First AUG - Protein Synthesis
Eukaryotic Translation and the First AUG
The sequences preceding the initiation codons in
eukaryotic messengers do not contain significant regions of complementarity to
the 18S RNA from the smaller ribosomal subunit. Additionally, translation
almost always begins on these RNAs at the first AUG codon. In bacteria, a
number of AUG triplets can precede the actual initiation codon. Trans-lation of
most eukaryotic messengers begins with the binding of a cap-recognizing protein
to the 5’ end of the mRNA. The translation efficiency of most, but not all,
messengers in eukaryotic systems is much higher when the messenger contains the
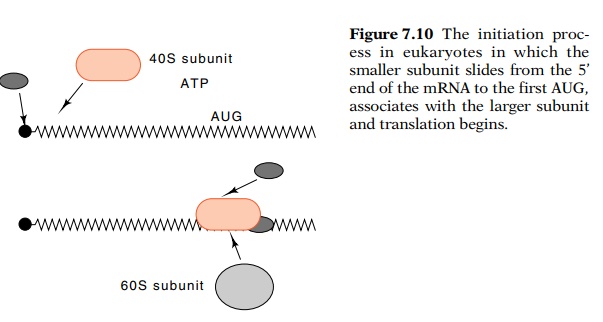
cap structure discussed. Additional proteins bind followed by the 40S ribosomal
subunit. With the consumption of ATP, the complex moves down the RNA to the
first AUG, at which point another protein binds to the complex and finally the
60S ribosomal subunit binds (Fig. 7.10). Trans-lation begins from this point.
The preinitiation complex can open and slide right through moderately stable
regions of base paired RNA to reach the first AUG of the messenger. This is
unlike the prokaryotic translation machinery, which has difficulty reaching an
initiation codon that is buried in extraneous secondary structure of the mRNA.
In special situations when only a low level of protein is required, translation begins not at the first AUG, but at a later one. Similarly, an AUG codon immediately followed by a termination
signal and then another AUG codon, also produces a low translation efficiency.
The need for a mechanism that can deposit the
translation machinery at the initiation codon despite the existence of
secondary structure in the mRNA arises from the eukaryotic translation pathway.
In eukary-otes, the RNA is synthesized, spliced, transported to the cytoplasm
and then translated. Undoubtedly, regions of secondary structure obscuring a
potential ribosome-binding site would exist on many species of mes-sengers. To
circumvent this problem, the translation apparatus recog-nizes the capped 5’
end, which cannot be involved in base pairing. After binding, the apparatus
slides down the mRNA until it reaches a starting AUG. In contrast, prokaryotes
lack a need for binding and sliding since ribosomes attach to a ribosome
recognition sequence on mRNA as soon as it protrudes from the RNA polymerase.
Thus, prokaryotic mRNA has little opportunity to fold and hide the start region
of a protein.
Related Topics