Chapter: Genetics and Molecular Biology: Protein Synthesis
Experimental Support for the Shine-Dalgarno Hypothesis - Protein Synthesis
Experimental Support for the Shine-Dalgarno
Hypothesis
Since the time of the original proposal by Shine
and Dalgarno, four lines of evidence have provided firm support for the idea
that the 3’ end of the 16S rRNA base pairs with a three- to seven-base stretch
of the mRNA lying ahead of the translation initiation site. The first line of
evidence is inhibition of an in vitro
protein synthesis system by an oligonucleotide. A polynucleotide of sequence
very similar to that which is found pre-
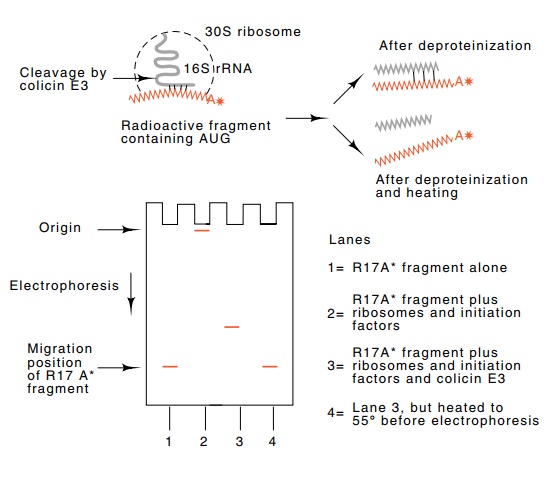
Figure
7.8 Base pairing between the 3’end of
16S rRNA and radioactivemessenger RNA. After incubation and before
electrophoresis, protein was denatured by addition of sodium dodecyl sulfate.
The mRNA barely entered the gel if it participated in forming an initiation
complex. Cutting the mRNA-16S rRNA complex with colicin E3 released mRNA
complexed to a fragment of 16S rRNA. Heating dissociated the mRNA fragment and
16S rRNA fragments.
ceding the AUG of many messengers inhibits mRNA
from binding to the ribosome. Presumably, this inhibition results from the
binding of the polynucleotides near the end of the 16S rRNA, thereby blocking
the ability of the ribosomes to bind properly to messenger.
The second line of evidence for the Shine-Dalgarno
proposal is direct physical evidence for base pairing between the messenger and
the 3’ end of 16S ribosomal RNA (Fig. 7.8). This experiment used a
bacteriocidal agent called colicin E3 which is released from some strains of
bacteria. E3 kills by inactivating the ribosomes in sensitive cells by cleaving
their 16S rRNA molecules 40 bases from the 3’ end. To test the ribosome-binding
site idea Jakes and Steitz first bound a fragment of phage R17 messenger in vitro to ribosomes in an initiation
complex and then cut the ribosomal RNA by the addition of colicin E3. They
demonstrated the presence of base pairing between the messenger and the
3’-terminal 40 bases of ribosomal RNA by co-electrophoresis of the fragment of
R17 RNA with the fragment of 16S rRNA. Repeating the experiment but preventing
formation of the mRNA-ribosome initiation complex pre-vented formation of the
hybrid between the two RNAs.
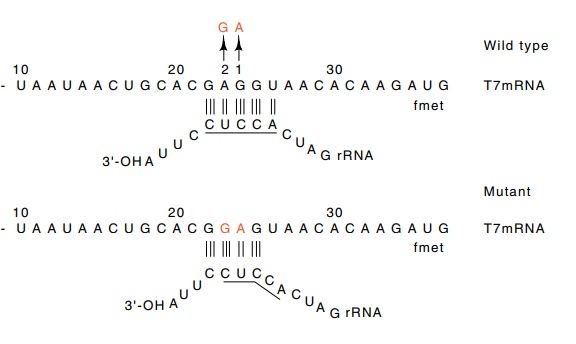
Genetics provides two additional arguments for the
utilization of the ribosome-binding site. A base change in phage T7 messenger
in the ribosome-binding site reduced the translation efficiency of the
messen-ger. The proof came with the isolation of a revertant that restored the
high translation efficiency. The revertant did not recreate the original
sequence; it created another ribosome-binding sequence in the mRNA, two
nucleotides further upstream (Fig. 7.9).
Figure 7.9 The base pairings possible between normal phage T7 messengerand 16S rRNA. Mutation 1 reduces the binding. Mutation 2, a revertant, restores the ability to base pair with the rRNA, but it pairs in a different position.
An elegant demonstration of pairing between mRNA and rRNA became possible with genetic engineering. One gene in E. coli was altered to possess a totally different ribosome-binding site. An addi-tional gene coding for the ribosomal 16S RNA was added to the same cells. Its recognition region near the 3’-OH end was changed to be complementary to the altered ribosome-binding site on the one gene. Only when both the altered gene and altered rRNA gene were present in cells was the protein from the altered gene synthesized. This proved beyond any doubt that the ribosome-binding sequence actually does base pair with a portion of the 16S rRNA.
Related Topics