Chapter: Genetics and Molecular Biology: Protein Synthesis
How Synthetases Identify the Correct tRNA Molecule
How Synthetases Identify the Correct tRNA
Molecule
A second
problem in specificity of protein synthesis is the selection of the tRNA
molecule by the synthetase. In principle this selection could be done by
reading the anticodon of the tRNA. A wide variety of experiments have revealed,
however, that only for tRNAMet is the anti-codon the sole
determinant of the charging specificity. For about half of the tRNAs, the
anticodon is involved in the recognition process, but it is not the sole
determinant. For the remaining half of the tRNAs, the anticodon is not involved
at all.
Two
extreme possibilities exist for the other charging specificity determinants.
They could be the identity of one or more nucleotides somewhere in the tRNA. On
the other hand, the charging specificity could be determined by part or all of
the overall structure of the tRNA molecule. Of course, this structure is
determined by the nucleotide sequence, but the structure as dictated by the
overall sequence may be more important than the chemical identity of just a
couple of amino or carboxyl groups. In view of the diversity of nature, it is
reasonable to expect different aminoacyl-tRNA synthetases will utilize
different struc-tural details in their identification of their cognate tRNA
molecules.
Just as
in the study of RNA splicing, the development of genetic engineering has
greatly accelerated the rate of progress in under-standing the specificity
determinants on tRNA molecules. This resulted from facilitating the synthesis in vitro of tRNA molecules of any
desired sequence. Such a synthesis utilizes the phage T7 RNA polymerase to
initiate transcription from a T7 promoter that can be placed near the end of a
DNA molecule (Fig. 7.5). Essentially any DNA sequence down-stream from the
promoter can be used so that any tRNA sequence can be synthesized. The RNA
molecules resulting from such reactions can
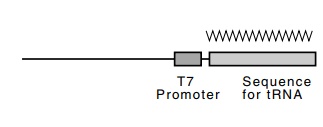
Figure
7.5 In vitrosynthesis ofan artificial tRNA
molecule using T7 RNA polymerase.
be aminoacylated and utilized in translation
despite the fact that they lack the specialized chemical modifications that are
found on tRNA molecules synthesized in
vivo. Apparently these modifications are not essential to the process of
protein synthesis and they exist more for fine-tuning.
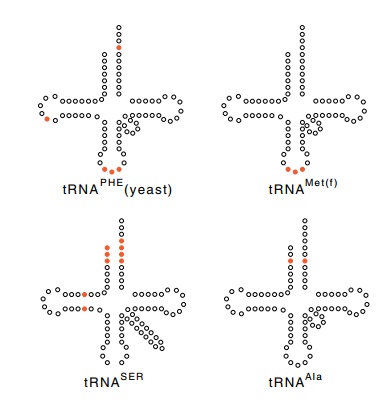
Figure 7.6 The positions ofnucleotides that determine the charging identity of sev-eral tRNAs.
Genetic engineering also enables us to alter the
gene encoding a tRNA molecule, reinsert the gene in a cell, and examine the in vivo charging and translation
properties of the altered molecule. The ability to be charged by alanine
synthetase is specified by the identity of just two nucleotides (Fig. 7.6).
This was determined by identifying the smallest common subset of nucleotide
changes that permitted the molecule to be charged with alanine. These proved to
be two nucleotides in the acceptor stem, a G and a U that form a non
Watson-Crick base pair. Providing these two nucleotides in any tRNA molecule
enables the molecule to be charged with alanine. The specificity determinants
of other tRNA molecules have been found to be three or more nucleotides
scattered around the molecule.
The structure of the crystallized glutamyl-tRNA
synthetase-tRNA complex permitted direct examination of the contacts between
the enzyme and the tRNA. These showed, as expected, that this enzyme read the
anticodon of the tRNA plus several nucleotides located elsewhere on the tRNA.
Related Topics