Chapter: Genetics and Molecular Biology: Protein Synthesis
Proportionality of Ribosome Levels and Growth Rates - Protein Synthesis
Proportionality of Ribosome Levels and Growth
Rates
The
dramatic ability of bacteria to grow with a wide variety of rates prompts the
question of how they manage to maintain balanced synthe-sis of their
macromolecules. In a study of this question, Schaechter, Maaløe, and Kjeldgaard
made the discovery that ribosomes are used atconstant efficiency, independent
of the cell growth rate. To appreciate their contribution fully, it will be
helpful first to examine a related question: what is the average rate of
protein synthesis per ribosome? As a first step we will estimate this value
using typical cellular parameters, then we will calculate this value more
carefully and include the effects of increase in the number of ribosomes during
a cell doubling time.
An
average bacterial cell with a doubling time of 50 minutes contains about 1 × 10-13 g protein and
about 10,000 ribosomes. Approximating the molecular weight of amino acids to be
100,
1 × 10-13 g protein is 1 × 10-13/102
= 10-15 moles amino acid;
10-15
moles amino acid is 6 ×
1023× 10-15 = 6 × 108
molecules;
104
ribosomes polymerize these 6 ×
108 amino acids in 50 min or 3 ×103sec.
Thus the
average rate of protein synthesis per ribosome is 6 × 108/3 × 103= 20
amino acids per second per ribosome. Compared to the typicalturnover number of
enzymes, greater than 1,000 per second, this is a low number. We have seen
already, however, the process of addition of a single amino acid to the growing
polypeptide chain is complex and involves many steps.
To
calculate accurately the rate of protein synthesis per ribosome during
steady-state growth, we must include the growth of the cells in the
calculation. This can be done in the following way. Define αr as the
relative rate of synthesis of
ribosomal protein, that is,
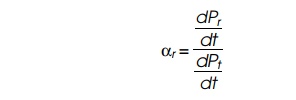
where Pr, is ribosomal protein and Pt is total protein. Let R(t) be the number of ribosomes in a
culture at time t. Since ribosome
number will
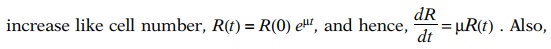
the rate of ribosome synthesis, dR/dt, equals the rate of ribosomal
protein synthesis in amino acids per unit time divided by the number of amino
acids in the protein of one ribosome, C:
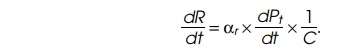
Then dPt/dt equals the number of
ribosomes times the average elonga-tion rate per ribosome, K. That is,
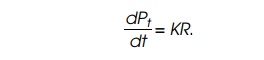
We have the following two expressions for dR/dt
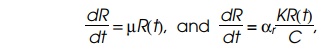
which
yields K =µC/αr, our
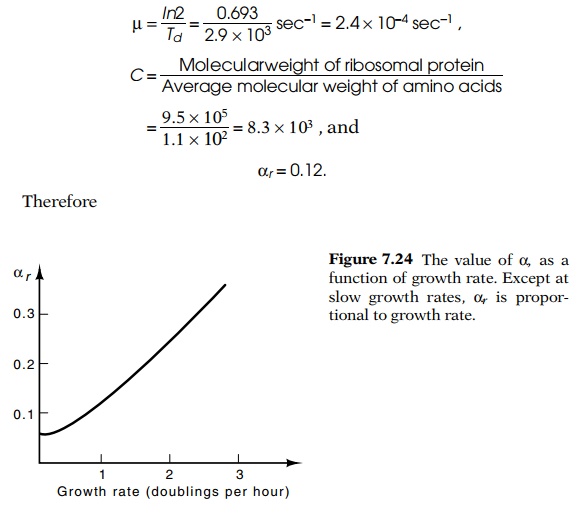
desired relation. Note that if αr is
roughly proportional to the growth rate, as has been found for bacteria except
at the slowest growth rates (Fig. 7.24), then the term µ/αr is a
constant and hence K, average
activity of a ribosome, is independent of the growth rate.
For E. coli
B/r growing at 37°, with a doubling time of 48 minutes,
= 17
aminoacidsper ribosomepersecond.
This
value is close to the elongation rate of polypeptides, showing that most
ribosomes in the bacterial cell are engaged in protein synthesis and are not
sitting idle.
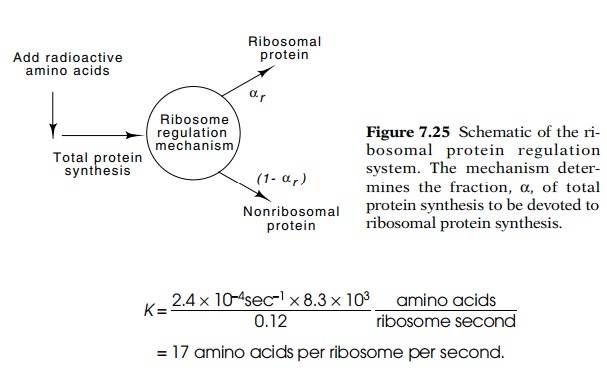
A
measurement of αr at any
time during cell growth can be accom-plished by adding a radioactive amino acid
to growing cells for a short interval (Fig. 7.25). Then an excess of the nonradioactive
form of the amino acid is added and cells are allowed to grow until all the
radioac-tive ribosomal proteins have been incorporated into mature ribosomes.
The value of αr, is the
fraction of radioactivity in ribosomal
protein compared to total radioactivity in all the cellular protein. This
fraction can be determined by separating ribosomal protein from all other
cellular protein by electrophoresis or sedimentation and measuring the
radioactivity in each sample.
Related Topics