Chapter: Biotechnology Applying the Genetic Revolution: Genomics and Gene Expression
Noncoding Components of the Human Genome
NONCODING
COMPONENTS OF THE HUMAN GENOME
Many noncoding DNA elements
have been identified in the genome of humans and other organisms. Many
scientists have referred to these regions as junk DNA because they are not expressed, either as proteins or RNA,
nor do they comprise regulatory regions associated with genes. However, some of
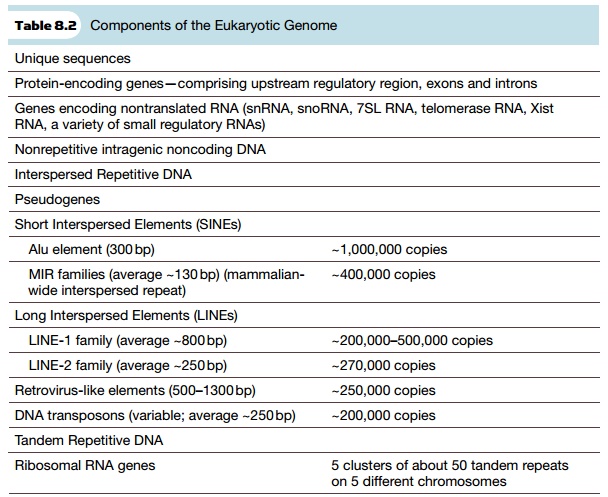
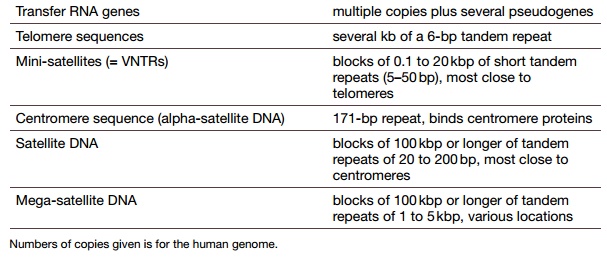
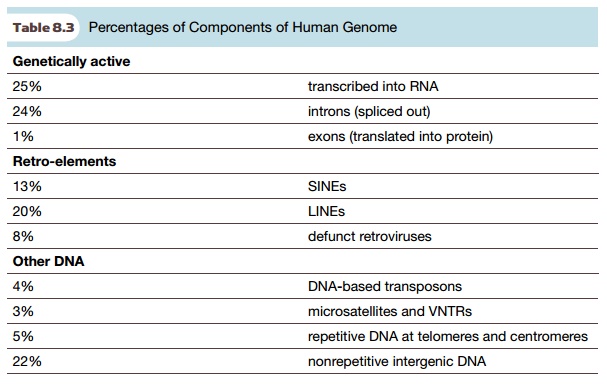
this DNA does have a functional role. Tables 8.2 and 8.3 list the major
components of the human genome and what percentage of each is present.
One common type of noncoding
DNA is the introns between genes. Most introns have no function, but there are
examples of whole genes being found within introns of a different gene. Introns
may also contain binding sites for transcription factors and therefore play a
role in gene regulation.
Another feature of the genome
is moderately repetitive sequences. Ribosomal RNA genes are found in great
numbers because many ribosomes are needed. These genes are considered
moderately repetitive elements, but of the coding variety. Noncoding repetitive
elements include LINE, for long interspersed element, which is
found in 200,000 to 500,000 copies (Fig. 8.12). These are retrovirus-like
elements that contain genes inside long terminal repeats (LTRs) similar to
retroviruses. They are autonomous, that is, the LINE is able to make copies of
itself and insert new copies into other sites in the genome. However, most
copies of LINEs are defective; only a few are still mobile and functional.
There are many different types of LINEs, the most common by far in mammals
being L1.

LINE retro-elements are transcribed into mRNA using the internal promoter. The mRNA goes to the cytoplasm, where ribosomes translate it, giving the proteins. One of these proteins, the reverse transcriptase/endonuclease dual function protein, then binds to the mRNA to form a ribonucleoprotein (RNP). This is transported back into the nucleus where the endonuclease portion nicks the DNA to generate a 3’-OH. Then the reverse transcriptase portion makes a DNA copy of the LINE that inserts into a new location in the genome. Cellular repair enzymes fill in the gaps to create duplicated sequences flanking both sides of the new LINE. When a LINE moves to a new location, it may disrupt an essential gene, which would prove fatal to the cell. Control of LINE movement is critical. Too much movement is disruptive and might destroy both the host cell and the LINEs it contains. Conversely, too little movement, and the LINE will fail to reproduce effectively. In humans, many LINEs are found in gene-poor, A/T-rich regions of the genome, suggesting that some mechanism exists to keep these elements from disrupting cellular function.
In addition to moderately
repetitive sequences, the human genome is filled with highly repetitive DNA.
The SINEs or short interspersed elements (see Fig. 8.12) are retro-elements like
the LINEs. The most common type of SINE is called the Alu element because an Alu
restriction enzyme site falls within the element. In the human genome, about
300,000 to 500,000 Alu elements exist. SINE elements are not able to move to
new locations in the genome without help from the LINE reverse transcriptase
and endonuclease protein. Unlike LINEs, SINEs are found in gene-rich regions of
the human genome, but they are shorter and often inert, so their presence does
not usually interfere with gene function.
Another type of highly
repetitive element found in the human genome is the minisatellite or VNTR. These were used in mapping the human genome,
and are scattered around the entire genome (see earlier discussion).
Related Topics