Chapter: Biotechnology Applying the Genetic Revolution: Genomics and Gene Expression
Gene Expression and Microarrays
GENE
EXPRESSION AND MICROARRAYS
As noted earlier, one of the
major issues in determining the correct number of genes is deciding whether or
not a sequence is really a gene. Measuring whether a gene is transcribed into
mRNA is the first step to deciding whether or not a presumed gene is real. Gene
expression was traditionally done on a single gene basis, but now, functional genomics studies gene
expression using the entire genome. Functional genomics encompasses the global
study of all the RNA transcribed from the genome—the transcriptome, all the proteins encoded by the genome—the proteome; and all the metabolic
pathways in the organism—the metabolome.
Because the entire human
genome contains only around 25,000 different genes, using microarray technology
to study gene expression is feasible. DNA
microarrays, or DNA chips,
contain thousands of different unique sequences bound to a solid support, such
as a glass slide. Microarrays are based on the principle of hybridization
between a “probe” and target molecules in the experimental sample. However, in
a microarray the probes are attached to the solid support and the experimental
sample is in solution. The microarray represents the genome of the organism
being tested and consists of sequences corresponding to each gene in the
organism.
To monitor gene expression,
RNA extracted from a cell sample is tested against a microarray. The
experimental RNA sample is usually fluorescently tagged. Hybridization of the
mRNA from the experimental sample to the DNA probes on the solid support
indicates whether or not a gene is expressed and to what degree. The level of
fluorescence at each point on the array correlates with the level of the
corresponding mRNA in the sample.
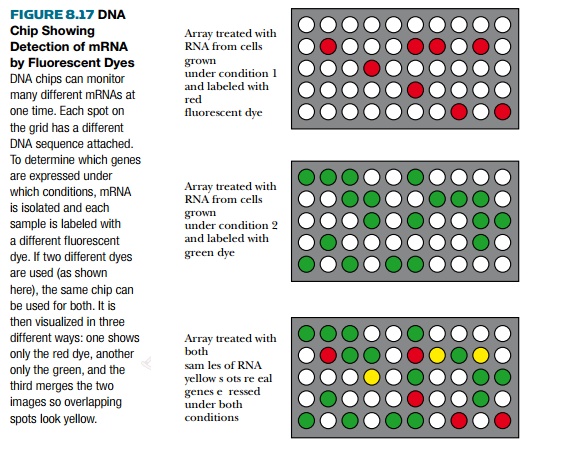
Microarrays can be used to
hybridize to RNA isolated from cells grown under a variety of experimental
conditions—for example, heat shock, cancer, or other disease states. The same
array can be hybridized to two or even more samples of RNA (control versus
experimental), thus comparing gene expression. For this comparison, each RNA
sample is labeled with a different fluorescent dye, for example, red and green.
If a particular RNA is present in only one of the samples,
the corresponding spot on the
microarray will be red or green (Fig. 8.17), whereas, if that particular RNA is
present in both experimental and control samples, the spot will be yellow
(i.e., red plus green). Modern arrays can accommodate thousands or millions of
different probes, allowing the entire genome for most organisms to be examined
at once. Some arrays are clustered so that all the genes involved in protein
synthesis are together, or the genes involved with heat shock are clustered.
The computer reads the color and fluorescence intensity for each of the spots
and carries out an analysis.
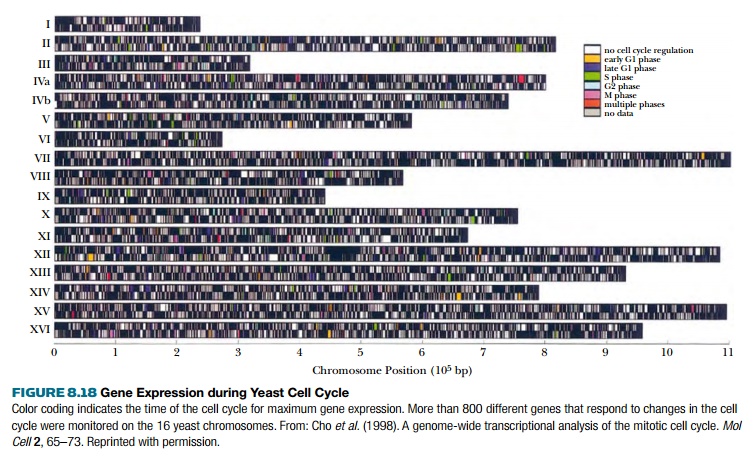
The results can provide a
global view of gene expression in different experimental conditions. This
powerful technique allows the researcher to analyze thousands to millions of
sequences at one time. For example, slides can be made with every named gene
from the yeast genome. These genes may be analyzed for expression at different
stages of the cell cycle. Thus, a culture of yeast can be synchronized and
arrested at different stages of the cell cycle by adding α factor or by using
mutant yeast that freeze at particular stages of mitosis. The gene expression
patterns for each stage are compared and compiled (Fig. 8.18).
Related Topics