Chapter: Biotechnology Applying the Genetic Revolution: Genomics and Gene Expression
From Pharmacology to Pharmacogenetics
FROM
PHARMACOLOGY TO PHARMACOGENETICS
Another field that has
undergone many changes due to the Human Genome Project is pharmacology, the
development of drugs to treat disease. Drug development has traditionally been
a hit-or-miss matter, with drug discovery often a by-product of other research.
Penicillin is one of the 20th century’s greatest discoveries, but was found by accident.
Alexander Fleming was growing Staphylococcus
bacteria and left his plates while on vacation. When he came back, mold had
contaminated the plates, and miraculously the staphylococci were not growing
close to the mold. The mold was evidently secreting something that stopped
bacterial growth.
Even Viagra was discovered by
accident. Scientists were trying to develop heart medications when they noticed
the “side effect.” One factor that makes drug development costs so high is that
many of the paths chosen lead to dead ends.
Another problem of drug
development is adverse drug reaction
(ADR). Adverse reactions may happen in some patients while others respond
well and are cured by the same drug. Most drugs are developed with the average
patient in mind, yet there is often a subset of people who react badly to the
drug. For example, many people are allergic to penicillin or other types of
antibiotics. Adverse drug reactions are a major cause of hospitalizations and
death. The differences in reaction to certain drugs often depend on the
person’s genetic makeup.
Pharmacogenomics is the study of all the genes that determine drug response in humans. More specifically, pharmacogenetics is the study of inherited differences in drug metabolism and response. The goal of these fields of study is to reduce the number of ADRs by determining the genetic makeup of the patient before offering a specific drug. The key to making a “genetic” diagnosis is the use of SNPs (see earlier discussion). Single changes in coding regions can often be correlated with adverse drug reactions. For example, if a certain subpopulation of people does not respond to a drug, then their DNA can be examined for a specific SNP that is absent in patients who do respond. Before the drug is given to any new patients, DNA from a blood sample can be tested for the presence of that SNP. Testing for SNPs can be done by microarray analysis (see later discussion) in the doctor’s office, thus reducing the number of office or hospital visits.
SNP analysis is also used to
screen for hereditary defects. Specifically, SNPs can be identified using a
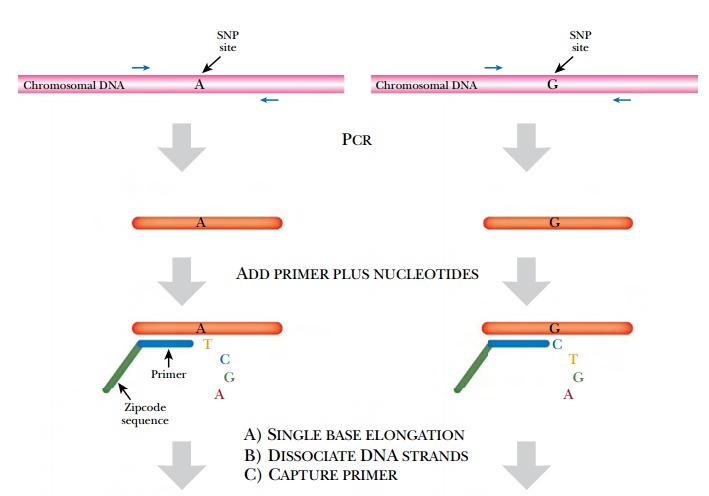
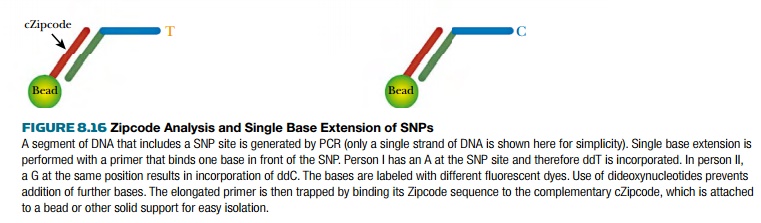
technique called Zipcode analysis (Fig. 8.16). Here many different SNPs can be
examined simultaneously. First, PCR is used to amplify the region containing
each different SNP being investigated. The PCR fragments could be sequenced in
full, but because SNPs differ by only one base, single base extension analysis
is done instead. For this, a primer is designed to anneal just one base pair
away from the SNP location. This primer also carries a “zipcode” region that is
used to identify this specific SNP, and each SNP has a different zipcode. After
the Zipcode primer anneals to the PCR fragments, DNA polymerase plus
fluorescently labeled dideoxynucleotides are added. This results in a single
base being added to the primer. (Note that dideoxynucleotides block chain
elongation, and so only one base can be added.) Each base is labeled with a
different fluorescent dye, allowing it to be identified. Next, beads linked to
complementary zipcode (cZipcode) sequences are added to grab the zipcoded
primers. The trapped Zipcode primer with the labeled nucleotide has a different
color based on which base was incorporated. The different colors can be sorted
and counted by FACS or fluorescent activated cell sorting.
One of the spin-offs from the
Human Genome Project is the Pharmacogenetics and Pharmacogenomics Knowledge
Base (PharmGKB; http://www.pharmgkb.org/). This records genes and mutations
that affect drug response. Consider asthma, a condition where people overreact
to inhaled irritants by cutting airflow in and out of the lungs. The muscle
cells around the bronchial tubes constrict, decreasing airflow. Albuterol is a
drug used to open the airways in people with asthma. This drug opens the
bronchial tubes by relaxing the muscle cells. Albuterol affects the
beta2-adrenergic receptor, and mutations in this receptor alter the efficacy of
albuterol. A single nucleotide change that replaces glycine at position 16 with
arginine gives a receptor protein with a better response to albuterol. Whether
a patient has this SNP will determine whether or not albuterol will be
effective.
Another key area of
pharmacogenetics concerns the cytochrome P450 family of enzymes. These play a
role in the oxidative degradation of many foreign molecules, including many
pharmaceuticals. The CYP2D6 isoenzyme oxidizes drugs of the tricyclic
antidepressant class, and different alleles of this enzyme affect how well a
person metabolizes these drugs. Much as for albuterol, identifying which allele
a patient has will prevent overdosages or adverse reactions. As time goes on,
more medical treatments will be designed for the individual rather than the
average person.
Related Topics