Chapter: Biotechnology Applying the Genetic Revolution: Genomics and Gene Expression
Bioinformatics and Computer Analysis
BIOINFORMATICS
AND COMPUTER ANALYSIS
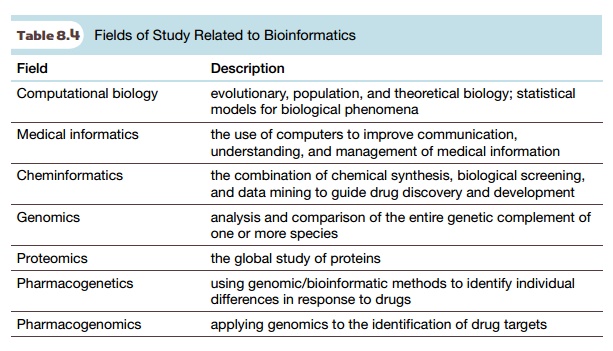
As noted before, the use of computers has revolutionized the way in which genetic information has been gathered and analyzed. The term bioinformatics has been coined to describe the emerging scientific discipline of using computers to handle biological information. It encompasses a large number of fields (Table 8.4). Bioinformatics includes the storage, retrieval, and analysis of data about biomolecules. By far the greatest achievement of the bioinformatics revolution has been the sequencing of the human genome. The term bioinformatics is now used to include analyses associated with DNA microarrays (see later discussion) and assessing the function of genomes.
Because bioinformatics is so
widely used, it is important to make genomic data available to researchers. The data from the Human Genome
Project are available on the Internet through the National Center of Biotechnology
website (http://www.ncbi.nlm.nih.gov/). Some other websites that present sequence data are
listed next. At the NCBI home page you can explore the human genome many
different ways. Using Entrez Gene, a specific gene can be identified by name.
The record for each gene contains the gene name and description, its location,
a graphical representation of the introns and exons for all the protein
isoforms that are known, and a summary of all the information known about the
gene. Additionally, the various domains within the protein, such as actin
binding sites, are listed with links to explain the domain and its function.
Finally, genes and/or regions of DNA from other organisms that are homologous
to the gene are shown. The page also contains links to research papers on the
gene’s function.
Some Bioinformatics Websites:
■
GenBank and linked databases
❍
http://www.ncbi.nlm.nih.gov/Entrez/
❍
http://www.ncbi.nlm.nih.gov/mapview/
❍
http://www.ncbi.nlm.nih.gov/genome/guide/human/
■
Institute for Genomics Research (TIGR)
❍
http://www.tigr.org/tdb
■
Genome Database (GDB) (human genome)
❍
http://www.gdb.org
■
European Bioinformatics Institute (including EMBL and Swissprot)
❍
http://www.ebi.ac.uk/
■
Flybase (Drosophila
genome)
❍
http://flybase.bio.indiana.edu:82
■
RCSB Protein Data Bank
❍
http://www.rcsb.org/pdb/
■
PIR Protein Information Resource (PIR)
http://www-nbrf.georgetown.edu/pir/searchdb.html
The program Map Viewer
(http://www.ncbi.nlm.nih.gov/mapview/) is used to browse the human genome
without any particular gene in mind. For example, individual chromosomes can be
explored via a graphical interface that allows you to zoom in and out of
various regions. Another genome browser
can be found at http://www.ensembl.org.
The amount of information
generated by the Human Genome Project is tremendous; therefore, understanding
this information without the use of computers is too difficult. Data mining refers to the use of
computer programs to search and interpret the data. Many bioinformatics researchers develop programs that search the
genomic data banks and sift, sort, and filter the raw sequence data. Data
mining programs often process information using the following steps:
1.
Selection of the data of interest.
2.
Preprocessing or “data cleansing.” Unnecessary information is
removed to avoid slowing or clogging the analysis.
3.
Transformation of the data into a format convenient for analysis.
4.
Extraction of patterns and relationships from the data.
5.
Interpretation and evaluation.
These programs can be designed to search for related sequences, determine areas of coding and noncoding DNA by looking at codon bias, or search for known consensus sequences, just to name a few applications. Searching for related sequences or similarity searches allows researchers to identify a potential function for a gene. If a gene of unknown function from humans is very similar to a characterized gene from flies, the two encoded proteins may have similar functions. This type of research is called comparative genomics. More than one gene can be compared. For example, entire pathways are often similar in different species. For example, human insulin attaches to a receptor on the cell surface and controls gene transcription via several intracellular proteins. Remarkably, very similar insulin signaling proteins are found in the roundworm Caenorhabditis elegans.
Difficulties may arise if scientists only use comparative genomics to study a new protein. Sometimes sequences that are similar have radically different roles as similar proteins may evolve new functions. Thus sequence similarity does not always imply functional similarity. Finally, the databases themselves are not perfect and may contain mistakes that are misleading. Comparative genomics must be complemented with other studies to reliably assign a role to a novel protein.
Other programs determine coding and noncoding areas of the genome by looking at codon bias. Identifying coding regions is critical for finding genes and can be accomplished by looking at the wobble position (third base) of the codon. Although a particular amino acid is often encoded by multiple codons, some codons are used preferentially. This codon bias varies from one organism to another. Most of the tRNAs for a particular amino acid will recognize the favored codon(s). For example, Escherichia coli genes preferentially use CGA, CGU, CGC, and CGG for the amino acid arginine, but rarely use AGA or AGG. Very few tRNAs for arginine are produced that recognize the AGA and AGG codons. Codon bias will thus be evident in regions that encode proteins, but in noncoding regions, the wobble position will not maintain this bias. Thus a potential gene in E. coli would contain relatively few AGA and AGG codons.
Finally, programs that
identify consensus sequences allow researchers to find various signatures or
motifs associated with particular functions. For example, a site that binds to
ATP has specific amino acids in specific locations. These sequences in an
unknown protein may help identify one of its functions. Other motifs include
actin binding domains, which indicate that the protein binds to the
cytoskeleton, or protease cleavage sites that suggest the protein is subject to
intracellular modification by proteases. Any potential motif in the sequence
must be confirmed experimentally. For example, a protein with an ATP binding
site signature must be shown to bind ATP experimentally. Thus, sequence
analysis provides a basis for further experiments.
Related Topics