Chapter: Biotechnology Applying the Genetic Revolution: Genomics and Gene Expression
Hybridization on DNA Microarrays
HYBRIDIZATION
ON DNA MICROARRAYS
Hybridization on a microarray
is similar to the hybridization of DNA during other hybridization experiments,
such as Southern blots, Northern blots, or dot blots. All these techniques rely
on the complementary nature of double-stranded DNA. When two complementary
strands of DNA are near each other, the bases match up with their complement,
that is, thymine with adenine, and guanine with cytosine. On a DNA microarray,
hybridization is affected by the same parameters as in these other techniques.
How the DNA is attached to
the slide can affect how well the probe DNA and target DNA hybridize,
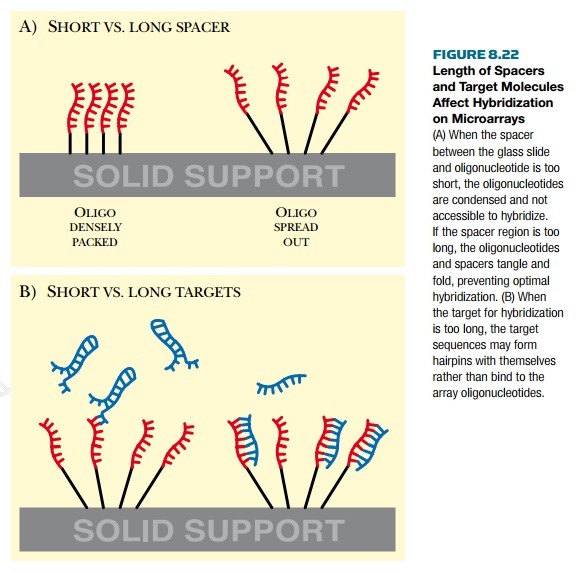
especially for oligonucleotide microarrays (Fig. 8.22). The short length of
oligonucleotides requires that the entire piece be accessible to hybridize. The
length of the spacer between the oligonucleotides and the glass slide optimizes
hybridization. An oligonucleotide attached with a short spacer has many of its
initial nucleotides too close to the glass and inaccessible to incoming RNA or
DNA. Oligonucleotides with longer spacers may fold back and tangle up;
therefore, again the sequence is inaccessible for hybridization.
Oligonucleotides attached with medium-sized spacers are far enough from the glass,
but not so far as to get tangled. Thus medium-sized spacers give the best
accessibility for hybridization.
Hybridization of two lengths
of DNA (or RNA with DNA) requires certain sequence features. One important
property is the relative number of A:T base pairs versus G:C base pairs.
Because G:C base pairs have three hydrogen bonds holding them together, it
takes more energy to dissolve the bonds. A:T base pairs have only two hydrogen
bonds and require less energy. Thus more GC base pairs give stronger
hybridization. If the sequence has many A:T base pairs, the duplex may form
slowly and be less stable. Another important consideration is secondary
structure. If the probe sequence can form a hairpin structure, it will
hybridize poorly with the target. If the probe has several mismatches relative
to the target, the duplex may not form efficiently. All these issues must be
addressed when making an oligonucleotide microarray. Computer programs are
available that identify suitable regions of genes with sequences that will
produce effective probes.
cDNA arrays are less prone to
the problems seen in oligonucleotide arrays. cDNAs are double- stranded, so
secondary structures such as hairpins are less likely to be a problem. During a
hybridization reaction, cDNA arrays must be denatured either with heat or
chemicals, making the probes single-stranded. Then the single-stranded RNA
samples are allowed to hybridize on the slide under conditions that promote
duplex RNA:cDNA without any mismatches.
Related Topics