Chapter: Biotechnology Applying the Genetic Revolution: Genomics and Gene Expression
Genetic Evolution
GENETIC
EVOLUTION
Genome sequencing has
expanded the field of molecular
phylogenetics, which is the study of evolutionary relatedness using DNA and
protein sequences. Comparing sequences from different organisms shows the
number of changes that have occurred over millions of years. All cellular
organisms, including bacteria, plants, and animals, have ribosomal RNA. These
sequences can be compared and the differences can be used to determine the
relatedness of different organisms. This system is less subjective than using
physical characters for taxonomy. The cladistic approach assumes that any
two organisms ultimately derive from the
same common ancestor (if we go far enough back) and that at some point bifurcation, or separation into two clades, occurred in their line of
descent. The difference between
the two organisms indicates
how long ago the split occurred. Taxonomy may be based on visible
characteristics—that is, the phenotype. This approach works well, at least to a
first approximation, in organisms with plenty of obvious features, such as
mammals and plants. But in organisms such as bacteria, the method falls apart.
However, molecular phylogenetics has opened the door to making family trees for
every organism.
When using molecular data to
study relatedness, it is essential that the sequences be correct and truly have
come from the organisms under study. This can be complicated in the human
genome because some sequences have been derived from other organisms, such as
viruses or bacteria. This problem applies to all organisms, to some extent. For
example, many bacterial genomes contain inserted bacteriophage genomes. Another
important point is to ensure that sequences being compared are truly
homologous, that is, they have all descended from one shared ancestral
sequence. When gene sequences are compared, they are aligned, so that the regions of highest similarity correspond (Fig.
8.13).
This type of alignment can
determine the relatedness of two or more proteins or genes. The relatedness can
be represented graphically by drawing phylogenetic
trees. The tree has various features: a root, nodes, and branches (Fig.
8.14). The root represents the overall common ancestor, and the branching
indicates the bifurcations or separations that occurred during evolution.

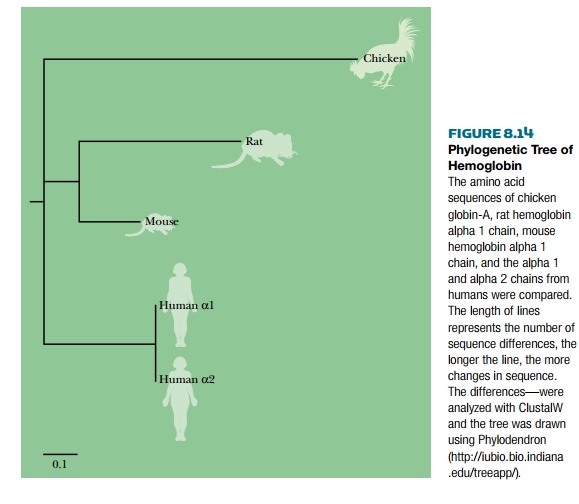
Individual nodes represent common ancestors between two subgroups of organisms. Branches represent clades, that is, groups of organisms with a common ancestor. The length of the branches indicates the number of sequence changes, so if the branches are short, the two organisms bifurcated relatively recently, and if the branches are long, the bifurcation occurred long ago.
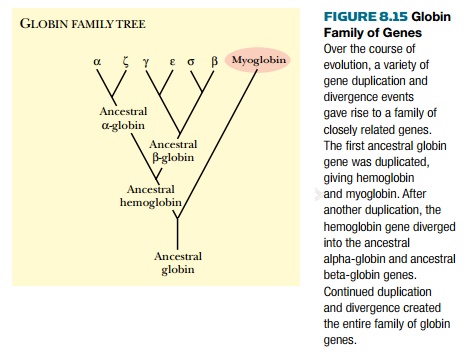
Based on alignments, genes
have been grouped into families,
groups of closely related genes that arose by successive duplication and
divergence. Gene superfamilies occur
when the functions of the various genes have steadily diverged until some are
hard to recognize. For example, the transporter superfamily encompasses many
proteins that transport molecules across biological membranes. This superfamily
has members that transport sugars into bacteria, transport water into human
cells, and even export antibiotics out of bacteria. They are found in almost
all organisms. Another gene superfamily is the globin family (Fig. 8.15). The
family includes myoglobin and hemoglobin from different organisms. These
proteins all carry oxygen bound to iron, but myoglobin is specific to muscle
cells whereas hemoglobin is specific to blood. The theory is that early in
evolution one gene for an ancestral globin existed. At some point this gene was
duplicated and the copies diverged so that one was specialized for blood and
the other for muscle. Hemoglobin itself also diverged later into different
forms, each used at various stages of development.
New genes may be generated
one at a time, but in addition, whole chromosomes or genomes may be duplicated.
In some organisms, particularly plants, genome duplications are relatively
stable and have occurred quite often. An example is the modern wheat plant. Its
ancestor was a typical diploid, but modern wheat used to make flour is
tetraploid. The wheat used to make pasta, durum wheat, is hexaploid and is
derived from three different ancestral plants. These varieties arose by natural
mutation and were exploited because of the higher protein content and better
yield.
The rate of mutation can vary
greatly between different genes. Although the human genome undergoes a steady
average rate of mutation, individual genes may mutate at different rates.
Essential proteins evolve or mutate more slowly than average. Conversely, the
less critical a gene is for survival, the more mutations can be tolerated and
the protein evolves more rapidly. Thus, the gene for cytochrome c, an essential component in the
electron transport chain, has incorporated only 6.7 changes per 100 amino acids
in 100 million years. In contrast, fibrinopeptides, which are involved in blood
clotting, have had 91 mutations per 100 amino acids in 100 million years. As
noted earlier, ribosomal RNA is useful to establish family trees for distantly
related organisms. It is found in every organism and is essential to survival;
therefore, it is slow to evolve.
What happens if a scientist
wants to classify organisms that are closely related? Essential gene sequences
do not provide enough genetic variation to differentiate such organisms.
Nonessential genes may help, but sometimes even these are too close. In such
cases, the wobble position of coding regions or even noncoding regions may be
used. As noted in Previews pagees, the wobble position is the third nucleotide
of a codon. The same amino acid is often encoded by several codons, which vary
only in this third base. Alterations at this position usually have no net
effect on protein function or structure and may occur between very closely
related species or between individuals of the same species.
Mitochondrial or chloroplast
genomes are also compared in order to determine the relatedness of organisms.
These accumulate mutations at a higher rate than the nuclear genomes in the same
organisms. The organelle genomes vary particularly in the noncoding regions.
One drawback to using organelle genomes is that mitochondria and chloroplasts
are inherited maternally and thus trace the evolutionary lineage only on the
maternal side.
Related Topics