Chapter: Biotechnology Applying the Genetic Revolution: Genomics and Gene Expression
Monitoring Gene Expression of Single Genes
MONITORING
GENE EXPRESSION OF SINGLE GENES
Although microarrays provide a
global view of gene expression, the results do not provide much specific
information on individual genes. Once candidate genes have been identified by
microarrays, these are analyzed individually. To determine the exact location
and/or level of expression, the gene of interest is genetically linked to a
reporter gene (see later discussion), creating a gene fusion. The regulatory region of the gene of interest is
isolated first. This segment is normally found upstream of the gene of interest
and includes sites for transcription factors to bind, plus various enhancer
elements. The coding sequence of the gene of interest is replaced with the
reporter gene, so that the regulatory elements now control the reporter gene
rather than the original gene of interest.
Reporter genes usually encode
enzymes whose activity is easy to assay. One of the most widely used reporters
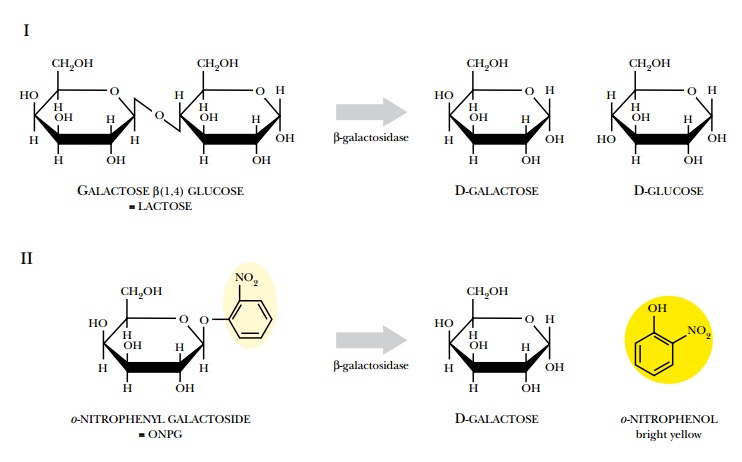
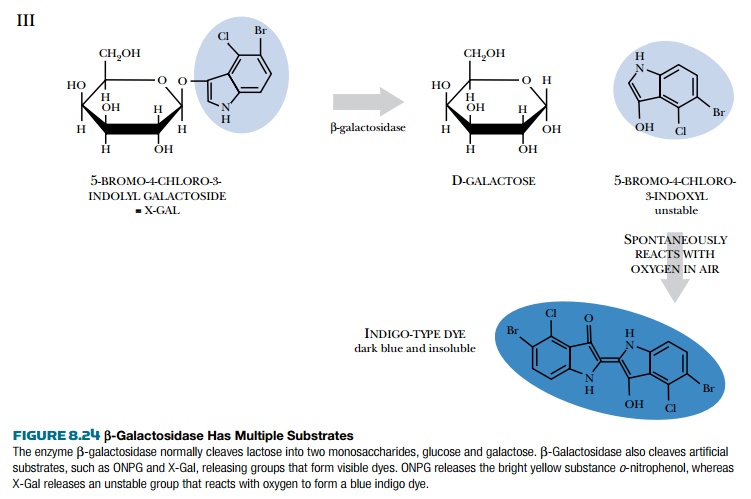
is the lacZ gene from E. coli, which encodes the enzyme b-galactosidase (Fig.
8.24). This enzyme splits disaccharide sugar molecules into their monomers, but
also cleaves various artificial substrates. When the substrate ONPG is cleaved,
one of the cleavage products forms a visible yellow dye. When X-Gal is cleaved
by β-galactosidase, one of the products reacts with oxygen to form a blue dye.
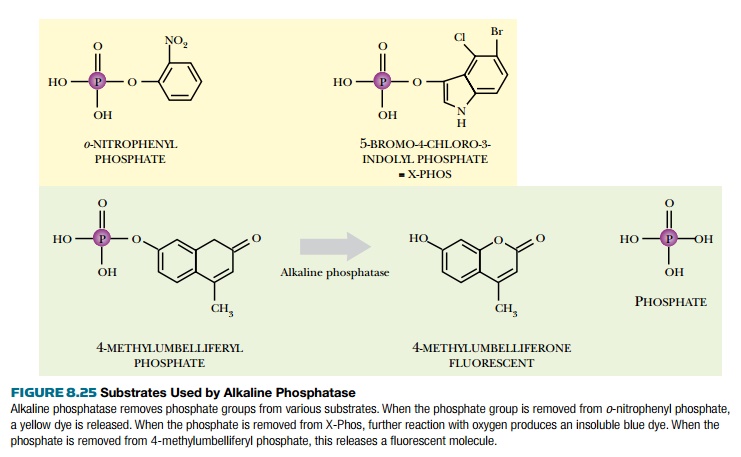
The phoA gene is another reporter gene that encodes alkaline phosphatase, which removes phosphate groups from many
different substrates (Fig. 8.25). Artificial substrates are designed so that
when the phosphate is removed, they either change color or fluoresce.
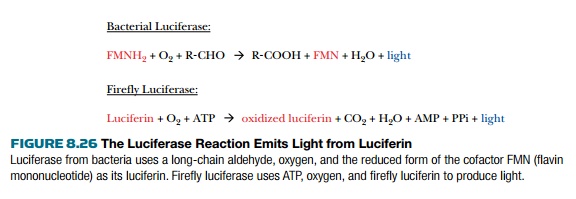
Another popular reporter gene is luciferase, which emits a pulse of visible light when the correct substrate, luciferin, is supplied (Fig. 8.26). Luciferase is an enzyme encoded by the lux gene in bacteria or the luc gene in fireflies. The two luciferases are not related and have different enzyme mechanisms. Both genes work well as reporter genes and have been cloned onto vectors that work in a variety of different organisms. Detecting the light emitted by luciferase is difficult because of its low levels and requires special equipment such as a luminometer or scintillation counter.
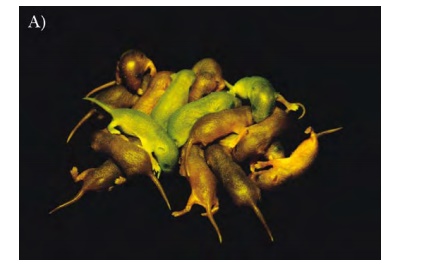
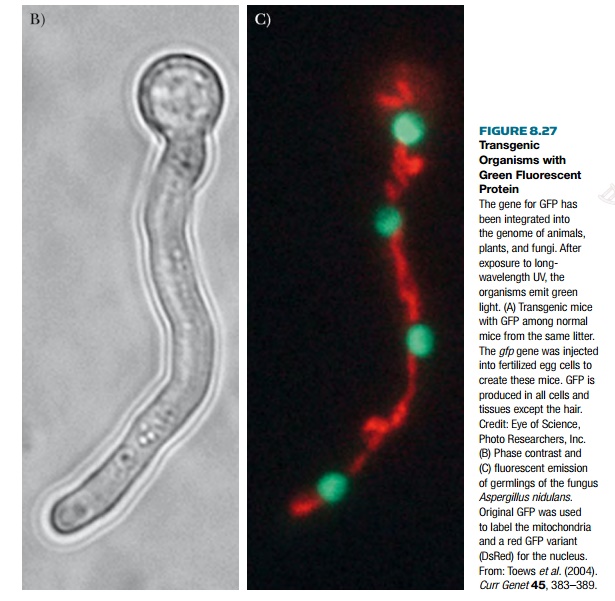
Another popular reporter
protein is green fluorescent protein
or GFP, which is not an enzyme (Fig.
8.27). This protein has natural fluorescence that does not require any cofactors
or substrates. Additionally, the fluorescence is active in living tissues, so
that when the protein is expressed the organism gains a green fluorescence.
This is especially noticeable when the organism is transparent like zebrafish
or Caenorhabditis elegans. GFP is
excited by long-wavelength UV light of 395 nm and then emits light at the green
wavelength of 510 nm. The original protein is from the jellyfish Aequorea victoria and is encoded by the
gene gfp. Many new variants of GFP
have been developed that emit light at different wavelengths, including red,
blue, and yellow. The main advantage of using GFP as reporter is the ability to
see expression in living tissues.
Other techniques are useful
to confirm data obtained with gene expression microarrays.
Differential display PCR is useful to compare mRNA expression patterns
from different tissue samples
or experimental conditions. Subtractive hybridization can also identify genes
that are expressed in one experimental condition and not elsewhere. Finally,
Northern blot analysis can monitor
expression levels of mRNA that vary in different experimental conditions.
Related Topics