Chapter: Biotechnology Applying the Genetic Revolution: Genomics and Gene Expression
DNA Accumulates Mutations over Time
DNA
ACCUMULATES MUTATIONS OVER TIME
The Human Genome Project has
opened the doors to improved analyses for many areas, including evolutionary biology. The sequence
features of the human genome arose over millions of years, as mutations, or
alterations of the genetic material, occurred and were passed on to successive
generations. During the course of human history many different events have
molded and sculpted our genetic history and resulted in our current genetic state.
Each individual has undergone some sort of genetic recombination and/or
mutation to become unique in physical and emotional constitution. Many genetic
mutations happen throughout all the cells of our bodies. Most of the defective
cells die and undergo apoptosis. When a mutation occurs in the somatic cells, the children or
offspring do not inherit the mutation, only when a mutation occurs in the germline or sex cells are the mutations
passed on to the next generation.
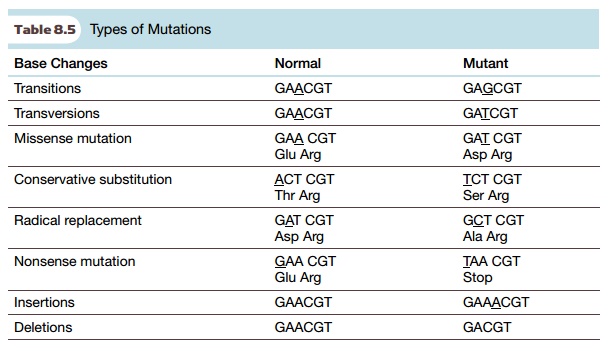
There are many different
types of mutations that cause genetic diversity (Table 8.5). The most common
are base substitutions, where one
nucleotide is exchanged for another. When a purine base is replaced by another
purine, or a pyrimidine is replaced by another pyrimidine, this is called a transition. If a pyrimidine is
exchanged for a purine, or vice versa, this is a transversion. These mutations create the SNPs used to make genomic
maps. Because different human individuals have variations every 1000 to 2000
bases, there are on average 2.5 million SNPs over the whole genome.
SNPs, or single base
substitutions, can fall anywhere in the genome, in either coding or noncoding
DNA. When the SNP falls within a gene, it may alter protein sequence and
function. When the base substitution alters one amino acid in a protein, the
mutation is called a missense mutation.
Some missense substitutions have little effect on protein structure or function
because one amino acid is replaced by another with similar properties. This is
known as a conservative substitution.
An example would be replacing threonine by serine, as these vary only slightly
in size but not in chemistry (both have an -OH group). Radical replacements, on the other hand, can alter the protein
function or structure because they
involve replacing amino acids with others that have a different chemistry.
For example, aspartic acid or serine are often involved in hydrogen bonding, and when either is replaced by a neutral amino acid like valine, the protein structure may become unstable. Sometimes missense substitutions create conditional mutations, where the protein will work under certain conditions but not others. A common conditional mutation is a temperature-sensitive mutation, where the mutation does not alter the protein function at the permissive temperature, but the protein is defective at the restrictive temperature. When base substitutions change a codon for an amino acid into a stop codon, this results in a truncated version of the original protein. These are nonsense mutations.
Besides base substitutions,
mutations may result in the insertion
or deletion of one or more bases. As
for single base substitutions, location is the key to what effect the mutation
will have. If the deletion or insertion of a few bases falls within a gene, it
may alter the reading frame of the protein, which will create random
polypeptide after the mutation. Often the altered reading frame creates a stop
codon, which truncates the protein. Large deletions may of course completely
remove all or part of a gene. Larger segments of DNA can undergo alterations
due to inversions, translocations, and duplications. Inversions occur when DNA
segments become inverted relative to the original sequence. Translocations
occur when DNA segments are moved to new locations. Duplications are when the
DNA segment is copied and then moved, resulting in two identical regions.
In some bacteria, reversible
inversions are deliberately used to cause phase
variation. Here, an enzyme called invertase
(strictly, DNA invertase) inverts a
segment of DNA involved in gene regulation. This alters the phenotype of the
bacteria. For example, Salmonella and
E. coli turn different surface
antigen proteins on or off by inverting a promoter segment.
In one orientation, the gene
is expressed and the corresponding antigen appears on the cell surface. In the
other orientation, the promoter is backward relative to the gene, which is
consequently not expressed. This allows the bacteria to vary their appearance
to the human immune system, making it easier to infect us.
To a first approximation,
mutations occur randomly throughout the genome. However, mutation hot spots are regions where mutations occur at much higher
frequencies. Mutations often occur
at methylated sites because methylated cytosine often loses an amino group,
turning it into thymine. DNA polymerase can also induce mutations during DNA
replication. Occasionally the proofreading ability of the polymerase fails and
single wrong bases are incorporated. More often, DNA polymerase undergoes strand slippage, when a segment of DNA
is highly repetitive. The result is either a duplication or deletion, depending
on the orientation of the slippage.
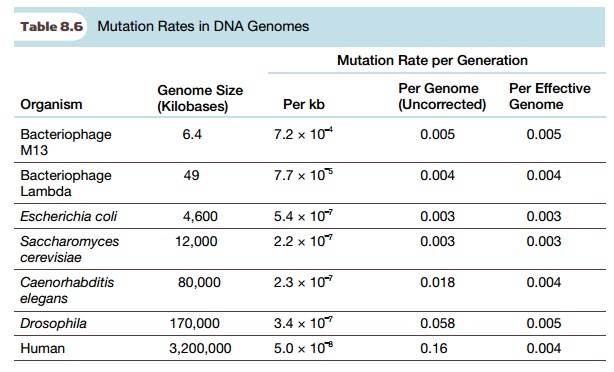
The rates at which mutations
occur help in understanding how mutations have affected the course of
evolution. The rate of mutation is low and depends on the organism and even the
particular gene being considered. Nonetheless, over long periods of time, many
mutations will occur. As Table 8.6 suggests, the rate of mutation is much lower
in genomes that are larger. In E. coli,
mutations occur at 5.4 × 10–7 per 1000
base pairs per generation, but in humans, mutations occur more than 10 times
more slowly, at only 5.0 × 10–8 per 1000
base pairs. However, when the mutation rate is corrected for effective genome
size (i.e., coding capacity rather than total DNA), it is approximately the
same for most organisms. This suggests that some mechanism must actively
control the mutation rate.
Related Topics