Chapter: Biotechnology Applying the Genetic Revolution: Genomics and Gene Expression
Making DNA Microarrays
MAKING DNA MICROARRAYS
There are two major types of DNA microarrays: one type contains cDNA fragments 600 to 2400 nucleotides in length, and the other uses oligonucleotides of 20 to 50 nucleotides in length. Each type of microarray is manufactured differently. When making a cDNA microarray, each of the different probes must be chosen independently and made by PCR or traditional cloning. Then all the DNA probes are spotted onto the slide. When making an oligonucleotide array, the oligonucleotide is synthesized directly on the slide.
cDNA Microarrays
The first step in making a cDNA microarray is to determine the numbers and types of probes to attach to the solid support. Since entire genomes have been sequenced for a variety of organisms, identifying potential genes is relatively easy. During the sequencing of these genomes, many cloned segments of DNA containing all or part of various genes were generated. Researchers can either obtain these clones or amplify genes from a sample of DNA using PCR. Each PCR product must be purified before attachment to the glass slide, so that all the extra nucleotides, Taq polymerase, and salts are removed and only pure DNA attaches to the slide. Pure cDNA samples can be used directly.

The next step is to create the chip using a microarray robot. A purified sample of each DNA is put into small wells arranged in a grid in microtiter plates. The size of the grid depends on the number of probes. If every predicted human gene is present once, approximately 25,000 different wells are needed. In practice, probes for each gene are attached more than once, in different areas of the chip, to provide several readings for each gene. A grid of pens or quills is dipped into the wells, one pen for each well, using a robotic arm. The pen tips are then touched to a glass slide, where a small drop of DNA is left behind. The robotic arm continues to manufacture spotted slides until the DNA in the well is used up. The use of a microarray robot makes each chip cheap and easy to produce. Finally, the DNA is crosslinked to the glass slide with ultraviolet light, which causes thymines in the DNA to crosslink to the glass. Figure 8.19 shows DNA on a microarray grid visualized by an atomic force microscope.
Newer technology has been developed to decrease the variation in size of spotted samples in microarrays. In newer cDNA microarrays, the samples are spotted onto a glass slide using inkjet printer technology. The cDNA samples are sucked into separate chambers of the inkjet printer head, and then spotted onto the glass slide much as ink is spotted onto paper in a printer. Inkjet technology prevents variations in size and quantity of cDNA in the sample spots. Special adaptors have been developed to prevent the inkjet sample channels from mixing, thus preventing cross-contamination.
Oligonucleotide Microarrays
Oligonucleotides are traditionally synthesized chemically on beads of controlled pore glass. Therefore, it is not too much of a logical leap to synthesize many different oligonucleotides side-by-side on a glass slide. The main differences between synthesizing single nucleotides on beads versus making arrays on glass slides is that the array has thousands of different oligonucleotides, and each must be synthesized in its proper location with a unique sequence. To accomplish this, photolithography and solid-phase DNA synthesis are combined (Fig. 8.20). Photolithography is a process used in making integrated circuits, where a mask makes a specific pattern of light on a solid surface. The light activates the surface it reaches, while the remaining surface remains inactivated.
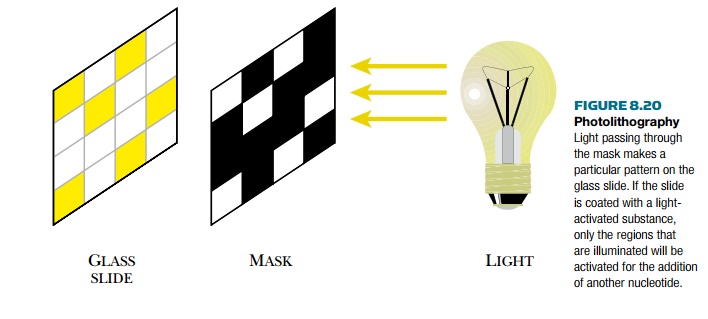
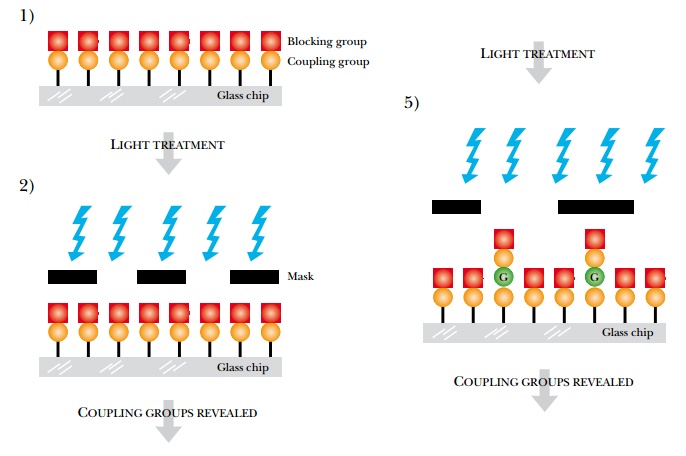
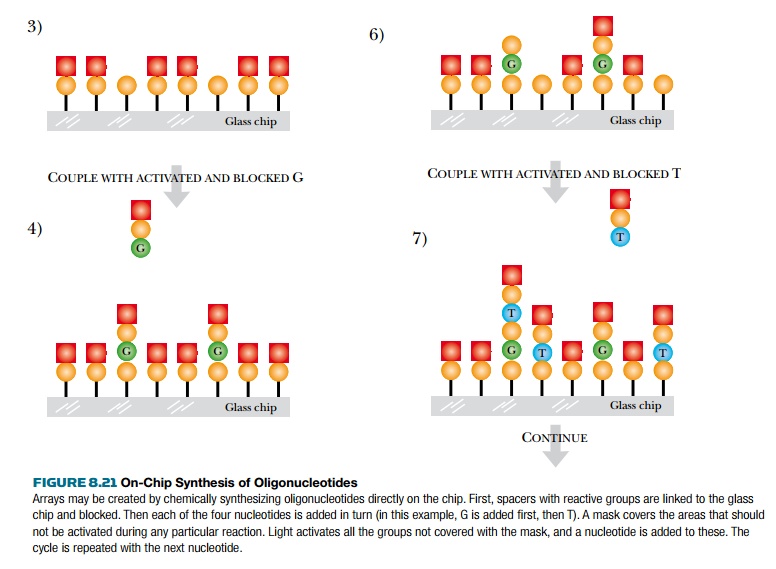
A glass slide is first covered with a spacer that ends in a reactive group. This is then covered with a photosensitive blocking group that can be removed by light. In each synthetic cycle, those sites where a particular nucleotide will be attached are illuminated to remove the blocking group. Each of the four nucleotides is added in turn. At each addition, a mask is aligned with the glass slide. Light passes through holes in the mask and activates the ends of those growing oligonucleotide chains that it illuminates. Much as in traditional chemical synthesis, each nucleotide has its 5′-OH protected. Thus after each addition, the end of the growing chain is blocked again. These protective groups are light activated, so at each step, a new mask is aligned with the slide, and light deprotects the appropriate nucleotides. The entire process continues for each nucleotide at each position on the glass slide. Making the masks is the key to this technology (Fig. 8.21).
Related Topics