Chapter: Biotechnology Applying the Genetic Revolution: RNA-Based Technologies
Engineering Allosteric Riboswitches and Ribozymes
ENGINEERING
ALLOSTERIC RIBOSWITCHES AND RIBOZYMES
Artificial or modified
ribozymes have enormous potential in medicine and biotechnology. The ability to
control the activity of a ribozyme would be very advantageous. If a ribozyme
were engineered to cleave mRNA that causes rampant growth of cancer cells,
controlling its action could prevent cancer from spreading. Additionally,
ribozymes could be engineered into genes used for gene therapy. By controlling
the ribozyme, the clinician could modulate when and where the gene is
expressed. Such control could be exerted by building riboswitches into the
ribozymes and then controlling self-destruction by the presence or absence of a
small effector molecule, as happens naturally in the glmS gene described
earlier.
In order to engineer a ribozyme to cleave only
in the presence of a certain effector molecule, a combination of modular design
and in vitro selection is used. Modular design takes various domains from
different ribozymes and merges them to create a new molecule. For example, the
catalytic core of a particular hammerhead ribozyme can be genetically linked to
the binding domain of another, changing the binding specificity of the original
ribozyme (Fig. 5.33A).
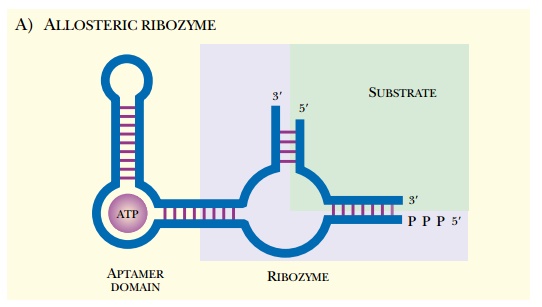
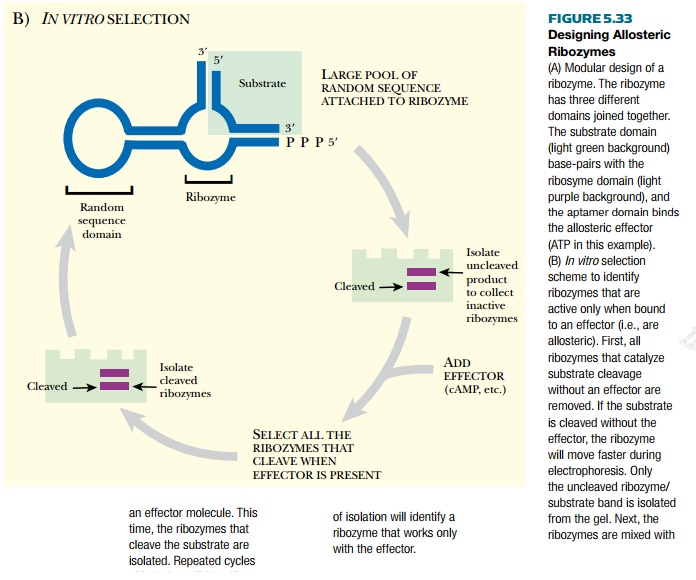
Artificial allosteric riboswitches have been
selected by combining the ribozyme catalytic core with a pool of many different
random sequences (see Fig. 5.33B). Some of the random sequences will, it is
hoped, have the ability to bind the chosen effector and thus represent a pool
of possible riboswitches. Some of the combinations will catalyze self-cleavage
or substrate cleavage without regulation, and these must be eliminated. If the
ribozyme construct cleaves itself, the products will move faster during
electrophoresis. Therefore, the pool of possible riboswitch/ribozymes is
electrophoresed, and the slower moving, uncleaved RNAs are isolated from the
gel. Next, the uncleaved ribozymes are mixed with the chosen effector and
incubated under cleavage-promoting conditions. This step is the positive
selection step, and any ribozyme that undergoes cleavage in the presence of the
effector is isolated. As before, the ribozymes are separated by gel
electrophoresis, but this time the cleaved (shorter and faster) molecules are
isolated. Cloning and sequencing of the isolated ribozyme constructs determines
the sequence of the riboswitch domain.
Some effectors that researchers have used to
control riboswitches include cyclic GMP, cyclic AMP, and cyclic CMP. Allosteric
ribozymes have been artificially created that respond not only to small organic
molecules such as cyclic AMP, but also to oligonucleotides, proteins, and even
metal ions.
Related Topics