Chapter: Biotechnology Applying the Genetic Revolution: RNA-Based Technologies
Riboswitches Are Controlled by Effector Molecules
RIBOSWITCHES
ARE CONTROLLED BY EFFECTOR MOLECULES
Transcription is controlled
primarily by protein factors. Nonetheless, in prokaryotes, conserved sequences
have been identified that control gene expression at the RNA level. These
sequences are an integral part of the messenger RNA molecules that they control
and are called riboswitches. Unlike
miRNAs or siRNAs, which work via base pairing, riboswitches bind small effector
molecules, such as nutrients or cAMP. The riboswitches work by alternating
between two different RNA secondary structures. In most cases, effector binding
terminates mRNA transcription prematurely or prevents mRNA translation.
Riboswitches are found in
several genes for biosynthetic enzymes. In E.
coli, the thiamine riboswitch is controlled by thiamine pyrophosphate, a
vitamin. When the vitamin is abundant, it binds to the TH1 box (i.e., a
riboswitch) close to the 5′ end of the mRNA, and
transcription of the mRNA is aborted. When the vitamin is absent, the mRNA is
transcribed and translated to give enzymes that make more thiamine. Similar
control occurs for riboflavin biosynthesis in Bacillus subtilis. The vitamin itself binds to the riboswitch
domain of the mRNA and controls whether or not the mRNA is expressed.
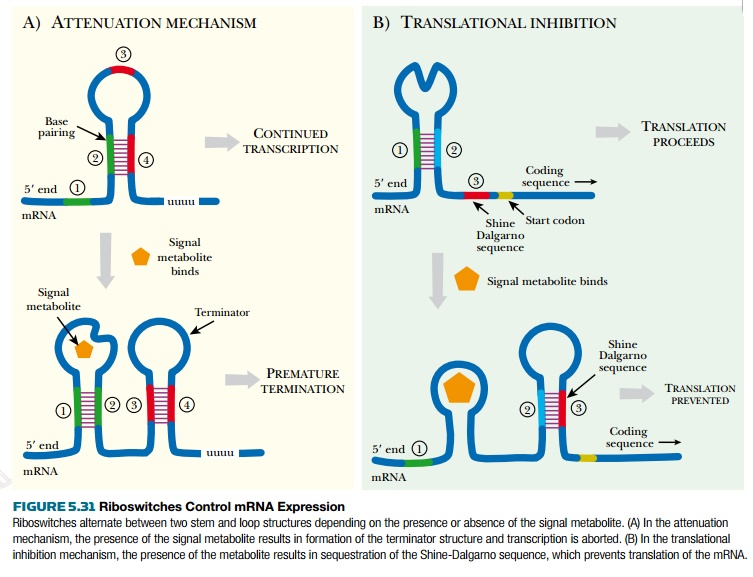
Riboswitches normally work by
changing the stem and loop structure of the mRNA transcript. In attenuation riboswitches, the effector
molecule binds to the mRNA as it is being transcribed. If the effector binds,
changes in structure create a terminator loop, which causes the transcriptional
machinery to fall off prematurely. The incomplete mRNA is degraded. When the
effector is in short supply, then the mRNA is transcribed to completion (Fig.
5.31A). Alternatively, some riboswitches work through translational inhibition.
Here the riboswitch controls whether or not protein translation occurs by
sequestering the Shine-Dalgarno sequence. When the effector molecule is
abundant, its binding changes the stem-loop structure so that the
Shine-Dalgarno sequence is not accessible to the ribosomes (see Fig. 5.31B).
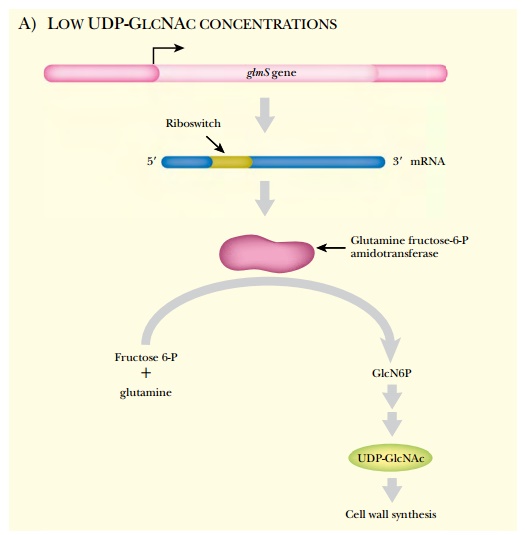
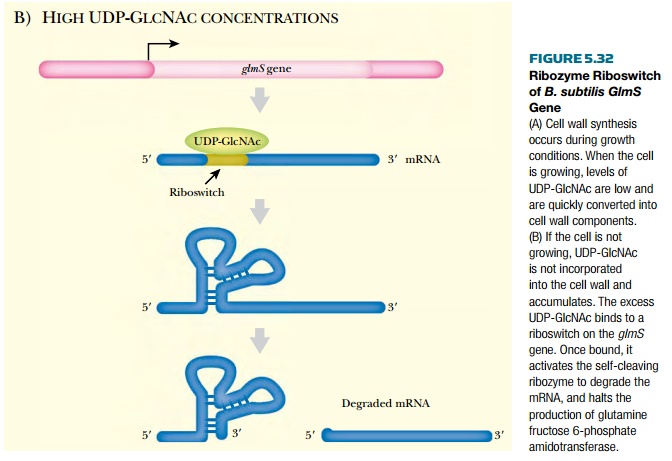
Recently, a novel riboswitch
was identified in Bacillus subtilis
that controls the expression of a biosynthetic gene (glmS) for a cell wall component (Fig. 5.32). As for other
riboswitches, a product of the biosynthetic pathway controls whether or not the
mRNA is expressed.
However, instead of hiding
the Shine-Dalgarno sequence or creating a terminator loop, the change in RNA
secondary structure creates a self-cleaving ribozyme. The glmS gene of B. subtilis codes
for the enzyme glutamine fructose 6-phosphate amidotransferase, which converts
fructose 6-phosphate plus glutamine into glucosamine 6-phosphate (GlcN6P). This
is further converted into a component of the cell wall, UDP-GlcNAc. When this
is abundant, it binds to glmS mRNA,
altering the secondary structure. The new structure functions as a ribozyme
that cuts the mRNA, preventing any further translation.
Other riboswitches have been
identified that respond directly to thermal stress. For example, the rpoH gene of E. coli is involved in the heat shock response. In addition to
other forms of regulation, the mRNA contains a thermosensor domain, which
controls the amount of translation. At normal temperatures, the thermosensor
has a stem-loop structure that prevents translation. When the heat increases,
the stem-loop structure falls apart and translation can occur.
Related Topics