Chapter: Biotechnology Applying the Genetic Revolution: RNA-Based Technologies
RNA Interference Uses Antisense Sequences to Inhibit Gene Expression
RNA
INTERFERENCE USES ANTISENSE SEQUENCES TO INHIBIT GENE EXPRESSION
RNA interference (RNAi) is a
recently discovered pathway for endogenous gene regulation where short
double-stranded RNA (dsRNA) segments trigger an enzyme complex to degrade a
target mRNA. In essence, the short dsRNA pieces decrease target protein
expression by degrading its complementary mRNA. RNAi was discovered in a
variety of different organisms, including plants, fungi, mammals, flies, and worms,
and consequently has multiple names. Different organisms have variations of the
same basic response. Mutations in the enzymes responsible for RNAi affect a
wide range of cellular processes. Some affect development of the organism;
others affect the ability to fend off viruses, particularly RNA viruses. In
still other cases, mutations affecting RNAi increase transposon movement,
suggesting that RNAi may also prevent transposon jumping. All these processes
rely on regulating mRNA translation or mRNA degradation.
RNAi is divided into two
different phases, the initiation phase and the effector phase. Initiation
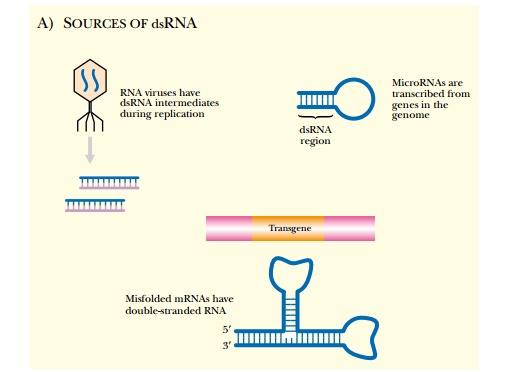
begins by the formation of dsRNA. The dsRNA can arise from three main sources.
First, externally infecting RNA viruses replicate through a dsRNA intermediate,
which can trigger RNAi. One theory suggests that the RNAi mechanism may have
evolved to combat these infecting viruses. Next, the organism’s own genomic DNA
contains sequences that code for microRNAs, which are specific mediators of
RNAi (see later discussion). Finally, dsRNA can be produced from aberrant
transcription of a genetically engineered gene (Fig. 5.10A). During the next
step of initiation, dsRNA binds to an endonuclease called Dicer, which cuts the dsRNA into small fragments about 21 to 23
nucleotides in length called short interfering RNAs (siRNAs; see Fig. 5.10B). Dicer is a dsRNA-dependent RNA
endonuclease that belongs to the
RNase III family. The siRNAs have a two-nucleotide overhang on the 3′ ends (characteristic of RNase III–type enzymes).
The 5′ ends are phosphorylated by a
kinase associated with Dicer, making the siRNAs competent for the next phase.
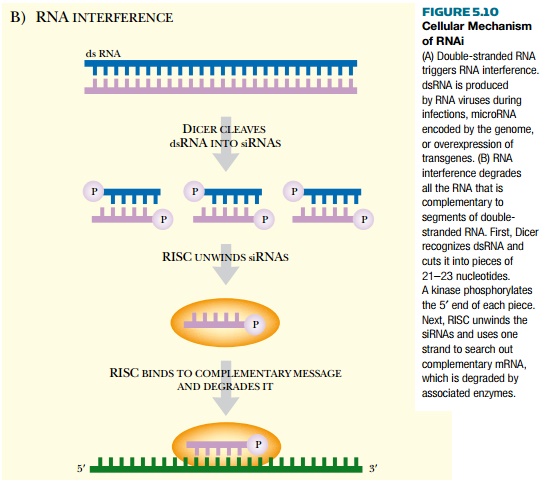
In the effector phase, Dicer transfers the siRNA to a ribonucleoprotein complex called the RNA-induced silencing complex (RISC). RISC is activated by the siRNAs and uses an RNA helicase to unwind the double-stranded fragments, making single strands. The antisense single-stranded siRNA is then kept as a guide to find complementary sequences in the cytoplasm. When RISC binds complementary sequences, these are first cleaved by an endonuclease within the RISC complex and are then further degraded by exonucleases in the cytosol. This destroys all of the mRNA that is complementary to the siRNA. Both Dicer and the RISC complex are dependent on ATP for energy. The antisense siRNA specifies which mRNA is targeted, ensuring that no nonspecific mRNAs are degraded.
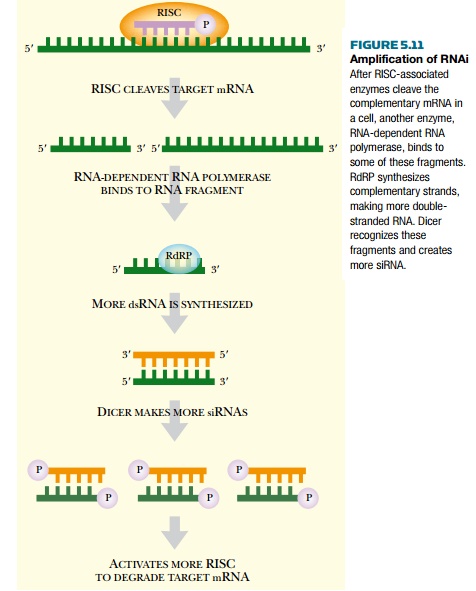
RNAi does not require many
molecules of siRNA. In fact, as few as 50 copies of siRNA may destroy the
entire cellular content of target mRNA. The ability to target so many mRNA
molecules with so few siRNA copies relies on amplification by the enzyme RNA-dependent RNA polymerase (RdRP), which creates dsRNA. RdRP uses the cleaved
target mRNA as template to
synthesize more dsRNA. Dicer recognizes the new dsRNA and cleaves it into more
siRNA, thus greatly increasing the number of siRNA molecules (Fig. 5.11).
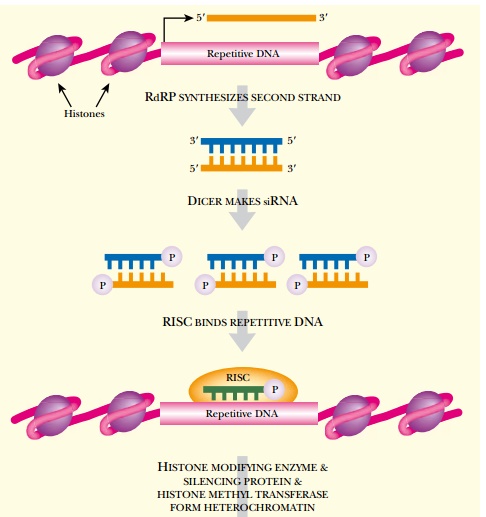
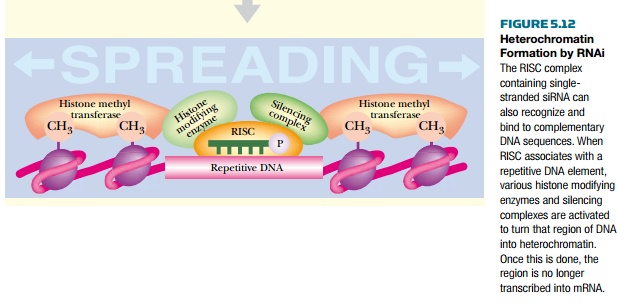
The final aspect of RNAi is
its ability to modulate DNA expression by converting copies of the target gene
into heterochromatin (Fig. 5.12). The siRNA can direct the heterochromatin-
forming enzymes and proteins to the target gene location. Once the open,
expressed DNA conformation is converted into heterochromatin, no more mRNA is
produced. Therefore, RNAi can repress gene expression permanently.
Related Topics