Chapter: Biotechnology Applying the Genetic Revolution: RNA-Based Technologies
Ribozymes Catalyze Cleavage and Ligation Reactions
RIBOZYMES
CATALYZE CLEAVAGE AND LIGATION REACTIONS
Ribozymes are RNA molecules
that bind to specific targets and catalyze enzymatic reactions. Some ribozymes
consist of RNA associated with proteins, but the RNA catalyzes the actual
reaction. Some ribozymes work like allosteric enzymes, that is, binding an
effector molecule alters the ribozyme structure so that the ribozyme becomes
competent to cleave its substrate. Ribozymes are naturally occurring, but
biotechnology research has started to exploit their unique characteristics for
medical and industrial applications.
There are eight known classes
of ribozymes at present, with the distinct possibility that many more will be
identified. Ribozymes are classified into large or small. The large ones range
in size from several hundred nucleotides to 3000 nucleotides in length. Large
ribozymes were the first identified, and the first of these were the group I
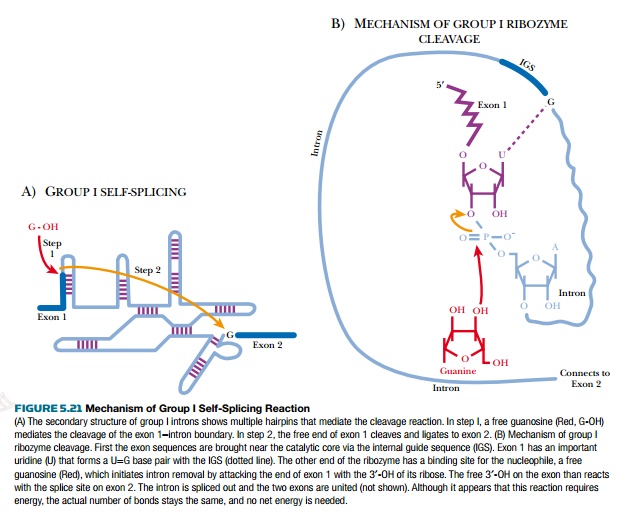
introns of Tetrahymena. These are
intron sequences found in pre-mRNA that are able to self-splice. They do not
use splicing factors such as U1, U2, U4/U6 snRNA (aka snurps). Group I introns
are common in fungal and plant mitochondria, in nuclear rRNA genes, in
chloroplast DNA, in bacteriophage and eukaryotic viruses, and in the tRNA of
chloroplasts and eubacteria. The important aspect of intron self-cleavage is
the RNA structure. RNA is a linear polymer, but because of base-pairing between
different regions, RNA also has a secondary structure. Multiple stem-loop
structures fold into different configurations leading to a three-dimensional
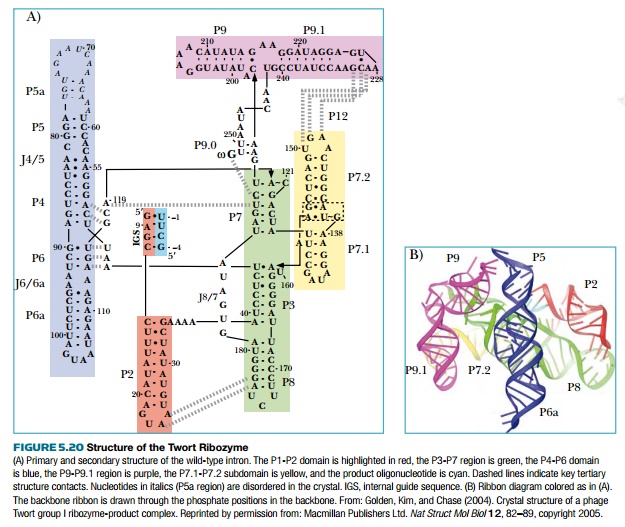
structure much like a protein (Fig. 5.20). The example shown is the second
group I intron within the orf142 gene
of bacteriophage Twort, which infects Staphylococcus
aureus. The three-dimensional structure of group I introns brings the two
exons close together, facilitating removal of the intron between them (Fig.
5.21).
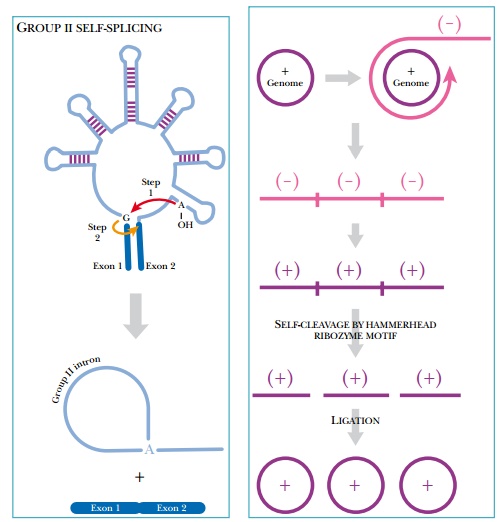

Group II introns are also
self-splicing sequences found within genes. They are less common than group I
introns, being found only in fungal and plant mitochondria, in plant chloroplasts,
in algae, in eubacteria, and in chloroplasts of Euglena gracilis. These introns do not self-splice in vitro and require far from
physiological conditions to work. The three-dimensional structure of the intron
creates these abnormal conditions in vivo,
affecting the microenvironment to create the correct ionic concentrations. The
3D structure of group II introns brings the two exons together, facilitating
intron removal and exon ligation (Fig. 5.22).
Another naturally occurring
large ribozyme is RNase P from bacteria. This is an RNA-protein complex, but
the RNA component is the catalytic entity. RNase P cleaves the 5′ end of pre-tRNA molecules. RNase P can act on
multiple substrates, unlike the group I and group II introns that naturally act
only on themselves.
Related Topics