Chapter: Biotechnology Applying the Genetic Revolution: RNA-Based Technologies
In Vitro Evolution and in Vitro Selection of Ribozymes
IN VITRO EVOLUTION AND IN VITRO SELECTION OF RIBOZYMES
It is also possible to
generate new ribozymes with novel enzymatic capabilities from large pools of
random RNA sequences. This is worthwhile because small natural ribozymes are
mostly limited to cleavage and ligation reactions. Using in vitro selection allows new ribozyme reactions to be identified
from random nucleotide sequences (Fig. 5.28).
For example, a ribozyme that
catalyzes the ligation of a particular sequence can be identified. This
approach begins by synthesizing a set of random oligonucleotide sequences.
However, these represent the pool of potential ribozymes rather than substrates
as seen in RNA SELEX. Each random sequence is flanked by two known sequences.
The 5′ end sequence is one
substrate for the desired ligation reaction. The 5′ end also has a terminal triphosphate to
energize ligation. The 3′ end has a sequence domain
that binds a chosen effector molecule. This allows the enzyme reaction to be
regulated. In addition, knowing the sequence at the two ends allows
amplification of the total construct by PCR.
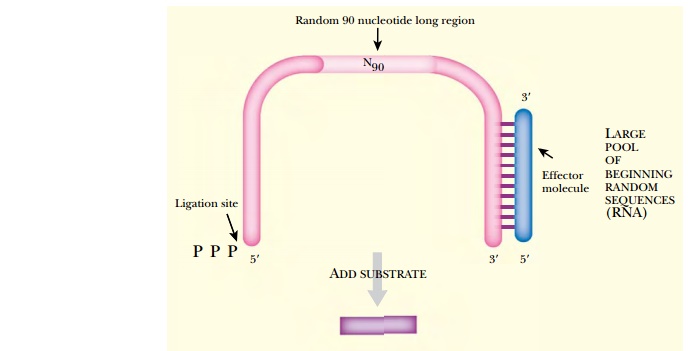
The other substrate for
ligation is mixed with the potential ribozymes and incubated in conditions that
favor ligation. If one of the random potential ribozyme sequences ligates the
substrate to its 5′ end, the resulting RNA
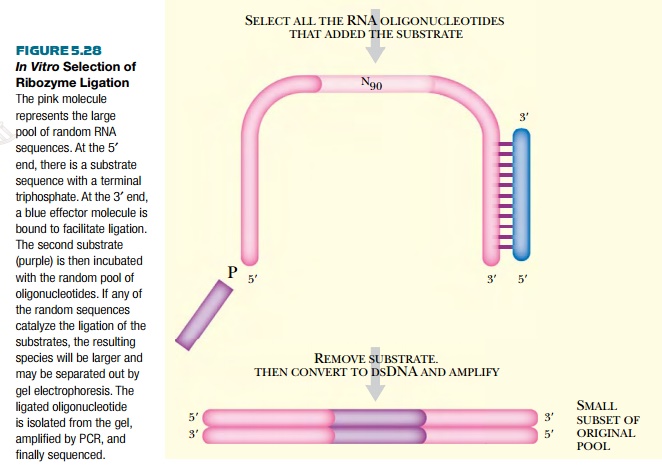
molecule (i.e., ribozyme plus ligation product) will run more slowly on an
agarose gel. The slower molecules are isolated from the gel. The ribozyme
suspect is then converted to DNA with reverse transcriptase. Finally, the DNA
is amplified with PCR using primers that match the 5′ end and 3′ end of the original RNA constructs.
In vitro evolution enhances in vitro selection
by adding a mutagenesis step after each cycle of selection (Fig. 5.29). This method begins with a pool of random
oligonucleotides, as before.
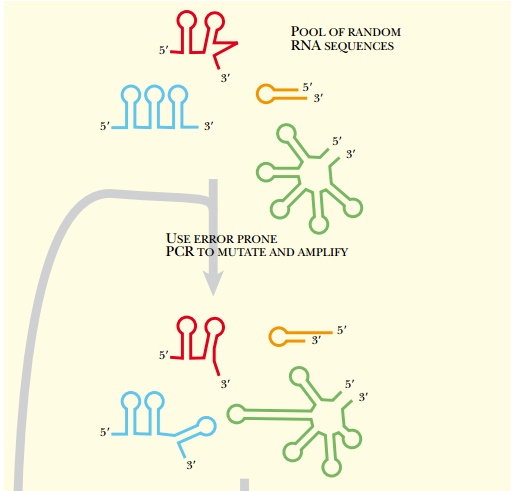
These sequences may be of any
length, provided they are long enough to carry out the desired reaction. The
pool of random sequences is then mutagenized. The most efficient method is to
use error-prone PCR to amplify the
initial pool of sequences. The pool both becomes larger and gains even more
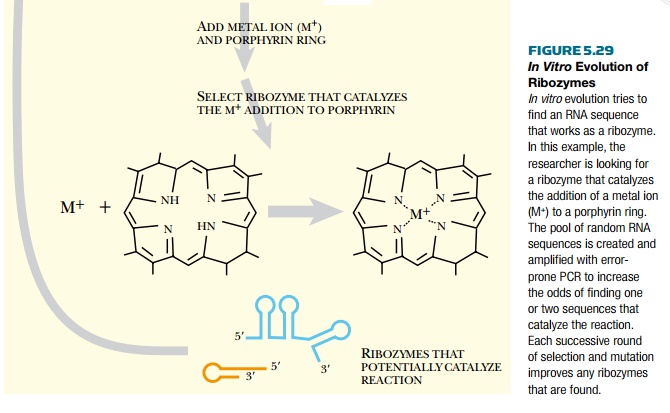
different sequences. The next step selects for the specific sequence that
carries out the desired reaction. For example, artificial ribozymes have been
evolved to add metal ions to mesoporphyrin IX (see Fig. 5.29). The mutagenesis
and selection steps can be repeated over and over to improve the ribozyme. Once
an efficient ribozyme is obtained, the sequence is determined after converting
the RNA into cDNA.
Artificial ribozymes have
been made to carry out nucleophilic attacks at various centers, including
phosphoryl, carbonyl, and alkyl halides. There is also an artificial ribozyme
that can isomerize a 10-member ring structure. In each of these cases, the
initial pools of oligonucleotides or ribozymes were selected for the ability to
carry out the specific reaction. In both in
vitro selection and in vitro
evolution, the key to success is the selection step. It must be stringent
enough that most of the nonfunctional RNA molecules are eliminated, but not so
stringent that ribozymes with weak activity are eliminated too early.
Related Topics