Chapter: Biotechnology Applying the Genetic Revolution: RNA-Based Technologies
Antisense RNA Modulates mRNA Expression
ANTISENSE RNA MODULATES MRNA EXPRESSION
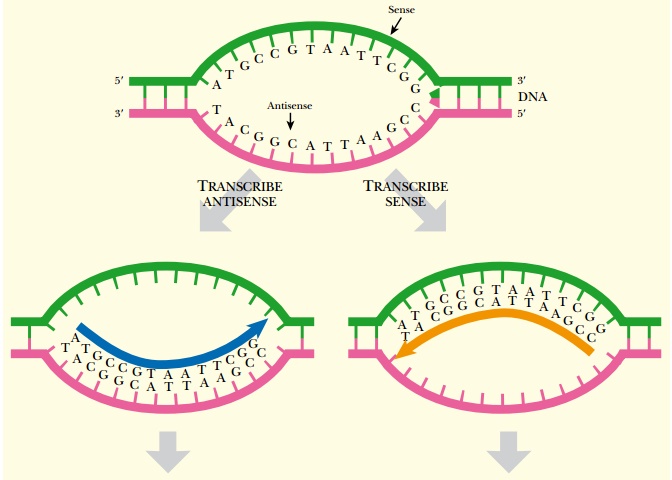
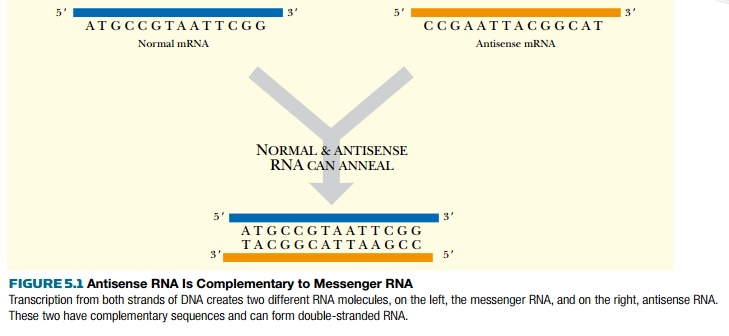
Antisense refers to the orientation of complementary strands during transcription. The two complementary strands of DNA are referred to as sense (= coding or plus) and antisense (= noncoding or minus;). Transcription occurs using the antisense strand as template, resulting in an mRNA that is identical in sequence to the sense strand (except for the replacement of uracil for thymine). Antisense RNA is synthesized using the sense strand as template; therefore, it has a sequence complementary to mRNA (Fig. 5.1).
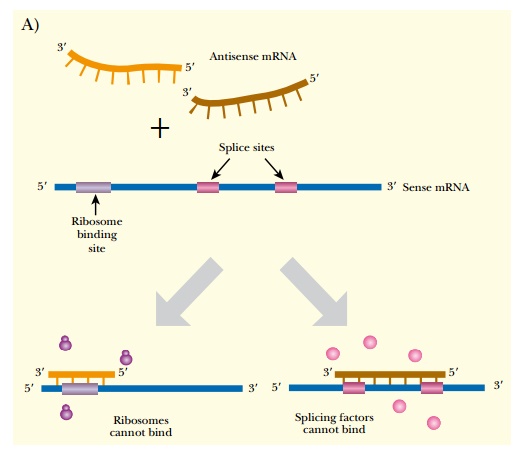
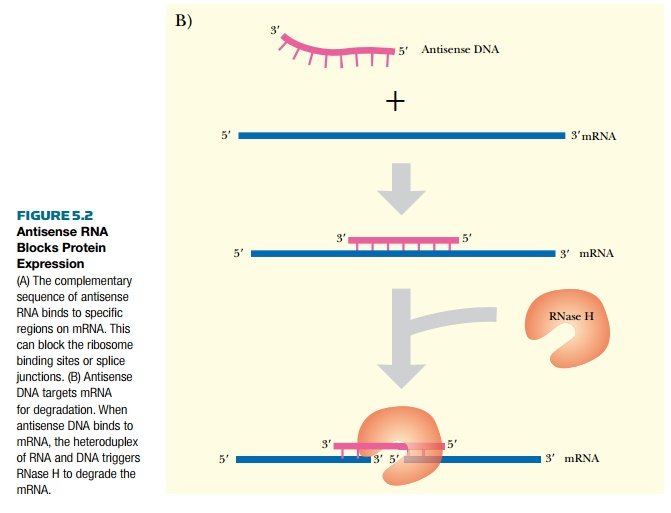
Antisense RNA is sometimes made in normal cells of many different organisms, including humans. Artificial antisense RNA is also made for manipulating gene expression in laboratory settings. When a cell has both the mRNA (i.e., the sense strand of RNA) plus a complementary antisense copy, the two single strands anneal to form double-stranded RNA. The duplex can either inhibit protein translation by blocking the ribosome binding site, or inhibit mRNA splicing by blocking a splice site (Fig. 5.2A). When antisense sequences are made in the laboratory, they are usually synthesized as DNA as this is more stable than RNA (see Chapter 4). In this case, the DNA:RNA duplex is digested by RNase H (see Fig. 5.2B). RNase H is a cellular enzyme that normally functions during replication. It recognizes and cleaves the RNA backbone of an DNA:RNA duplex, targeting the antisense DNA:mRNA duplex for further degradation. RNase H recognizes a 7-base-pair heteroduplex, so the region of homology between the antisense DNA and target mRNA need not be very long.
Antisense Controls a Variety of Biological Phenomena
Naturally occurring antisense genes have been found that control a variety of different processes. One example of natural antisense control is found inNeurospora. This fungus follows a strict schedule, based on circadian rhythms, and forms hyphae only at specific times during the day. Many mutants have been identified that do not follow such a strict standard of time. The genes affected by these mutations are regulators in the circadian rhythm ofNeurospora. One of the first circadian rhythm genes identified was frequency (frq).
Mutations in this gene change how often the fungus forms hyphae. The amount of frq mRNA fluctuates, with highest levels during the day and lowest at night. Interestingly, antisense frq RNA also cycles, but in reverse, with the lowest levels during the day and the highest during the night. Although the exact mechanism is uncertain, Neurospora that do not produce antisense frq RNA have disrupted circadian rhythms. In addition, both the antisense and sense mRNAs are induced by light and therefore respond directly to the environment to maintain the correct circadian rhythm.
Using antisense to regulate gene expression is so widespread in nature that scientists became curious how many potential antisense/sense partners exist in various genomes. Computer algorithms have been devised to search for sequences that could function as antisense. In the human genome, there are a predicted 1600 different partners. The most interesting finding in the antisense field is the realization that small noncoding regulatory RNAs calledmicroRNAs (miRNAs) inhibit gene expression through an antisense mechanism (see later discussion). Using computer searches, an additional 250 potential microRNAs have been identified in humans, but because these are only about 20 nucleotides long, identifying them conclusively by computer is very difficult.
EXAMPLES OF NATURAL ANTISENSE CONTROL
■ Control of ColE1 Plasmid Replication: RNAI and RNAII mRNA are sense/antisense partners that prevent DNA polymerase from initiating plasmid replication. The amount of antisense RNAI controls how often replication is initiated.
■ Control of X-Chromosome Inactivation: Antisense RNA binds to the Xist gene to prevent its expression and thus prevents X chromosome inactivation.
■ Control of Circadian Rhythm in Neurospora: The time of day controls when the fungus forms hyphae by regulating the antisense and sense mRNA for the frq gene. The antisense and sense mRNAs are controlled by light and dark cycles.
■ Iron Metabolism in Bacteria: FatB/RNAα are sense/antisense partners that control regulation of iron uptake in the fish pathogen Vibrio anguillarum. When iron is plentiful and iron scavenging is not necessary, higher amounts of RNAα prevent fatA and fatB expression. When iron is scarce, the bacteria need to get iron from the environment. RNAα mRNA is degraded and fatA and fatB are expressed so Vibrio can ingest iron.
■ Control of HIV-1 Gene Expression: Antisense env mRNA binds to the Rev Response Element (RRE) on env mRNA. When antisense blocks the RRE, Env protein is not produced. When antisense env mRNA is absent, Env protein is produced.
■ Control of RNA Editing: Antisense/sense loops are formed between complementary exon and intron sequences of the gene for the glutamate-gated ion channel in human brain. These loops are recognized by dsRNA-specific adenosine deaminase (DRADA), which converts adenosine to inosine by deamination. This alters the sequence of the final mRNA and hence of the protein. This reduces the permeability of the ion channel.
■ Alternate Splicing of Thyroid Hormone Receptor mRNA: Antisense RNA transcribed from the thyroid hormone locus inhibits splicing of the c-erbAα gene. Two alternately spliced transcripts give the authentic thyroid hormone receptor and a decoy receptor that does not bind thyroid hormone. These two forms modulate cellular responses to thyroid hormone.
■ Developmental Control of Basic Fibroblast Growth Factor (bFGF): Basic FGF controls various developmental and repair processes, including mesoderm induction, neurite outgrowth, differentiation, and angiogenesis (blood vessel development). Antisense bFGF mRNA binds to cellular bFGF mRNA during development of Xenopus laevis (African clawed frog), targeting the bFGF mRNA for degradation.
■ Control of Eukaryotic Transcription Factors: The transcription factor hypoxia-induced factor (HIF-1) is a basic helix-loop-helix dimeric protein that turns on genes associated with oxygen and glucose metabolism, including glucose transporters 1 and 3 and enzymes of the glycolytic pathway. Antisense mRNA to the α subunit mRNA controls the expression of the transcription factor. The level of antisense RNA is modulated by the amount of oxygen in the environment.
Related Topics