Chapter: Biotechnology Applying the Genetic Revolution: RNA-Based Technologies
Functional Screening with RNAi Libraries
FUNCTIONAL
SCREENING WITH RNAi LIBRARIES
RNAi has been used to study
the entire genome in model organisms. An RNAi library containing about 86% of
the predicted genes in the genome of C. elegans has been constructed in E.
coli. An RNAi library expresses each of the genes as dsRNA. E. coli carrying
any particular gene can be used to feed C. elegans, which triggers RNAi for the
gene of interest and allows the effect of removing each individual protein from
the worm to be assayed. Using this library, more than 900 genes have been
identified whose suppression kills the embryo or causes gross developmental
defects. Most of the proteins encoded by the RNAi library target mRNA that had
no known function before. These libraries demonstrate how RNAi can link a
particular gene to a specific cellular process. Similar experiments in
Drosophila have begun. An RNAi library of about 7200 conserved genes (about 91%
of the predicted genes in the genome) has been constructed. So far the library
has been used to identify proteins associated with cell growth and viability in
Drosophila cell lines, but the library can be applied to many different
processes.
In order to construct an RNAi
library, the genes from the organism of interest are isolated as cDNA clones
(Fig. 5.17). The cDNA clones are then amplified using PCR.
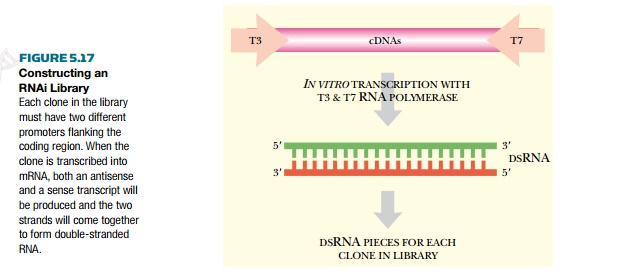
Because the genomes concerned
have been sequenced, PCR primers that are specific for each gene are designed
with special features. They are designed to add two different promoters at the
ends of each gene. For example, the 3′ end would have a T7 polymerase promoter,
and the 5′ end a T3 polymerase promoter. The PCR products are then cloned into
a suitable vector. When the vector is present in a cell with both T7 and T3 RNA
polymerase, both an antisense and a sense transcript are transcribed for each
of the clones. The two strands spontaneously anneal to form dsRNA, which then
activates RNAi.
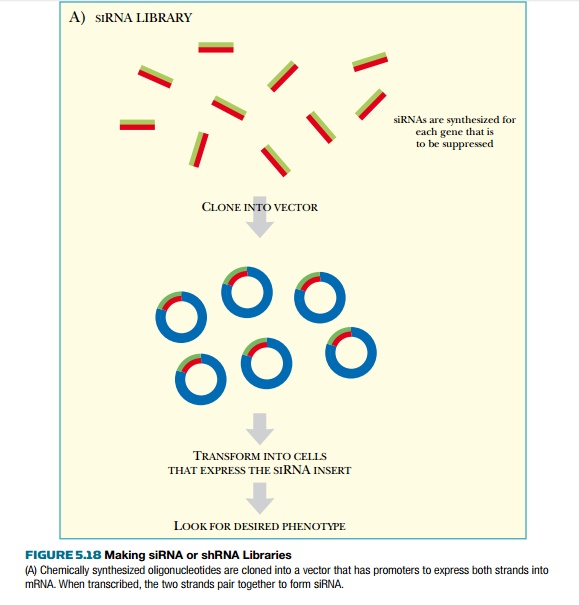
For mammalian systems, RNAi
libraries must be constructed with siRNAs or shRNAs rather than full-length
dsRNA, because the full-length dsRNAs are toxic. These RNAi libraries are made by
two different methods (Fig. 5.18). In the first method, short synthetic siRNAs
are chemically synthesized for each of the genes that will be represented in
the library. All of the siRNAs are pooled and cloned into the appropriate
vector. The vector must have the promoters and terminators needed to express
the siRNA inserts as double-stranded RNA. The promoters are usually two RNA
polymerase promoters that transcribe the sense and antisense strands of the
insert. As described earlier, these spontaneously anneal to make siRNA. The
vector must also contain other appropriate sequences, such as an origin of
replication for mammalian and bacterial cells, and antibiotic resistance genes
for selecting cells carrying the vector.
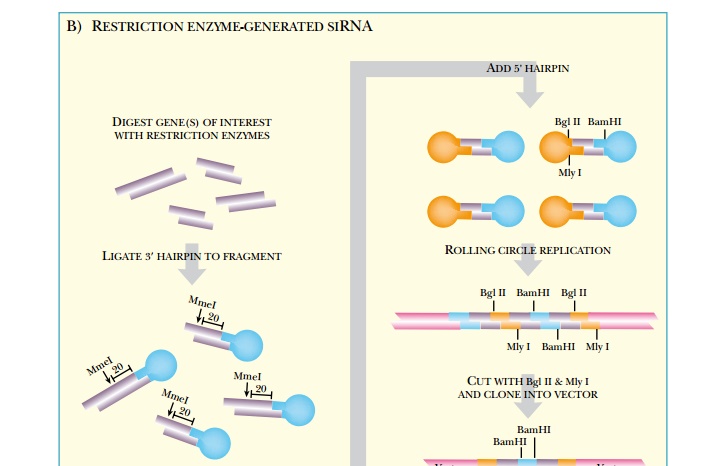
A second approach to
constructing a mammalian RNAi library involves the use of restriction enzymes
to generate small shRNA fragments. Restriction
enzyme-generated siRNA (REGS) libraries start with either single or
multiple genes that are going to be represented in the library (see Fig. 5.18B).
Whether a large or small sample of genes is used, the technique begins with
double-stranded cDNAs. The library is cut with restriction enzymes that cut
often and leave identical overhangs. This generates many different small
fragments for each gene. Because each of the fragments is a different size,
they must be trimmed to the same size. For this, the fragments are linked to a
3′ hairpin loop, which has a
site for the restriction enzyme MmeI.
This cuts precisely 20 nucleotides from the 3′ end of its recognition
sequence, thus making each fragment
the same length.
\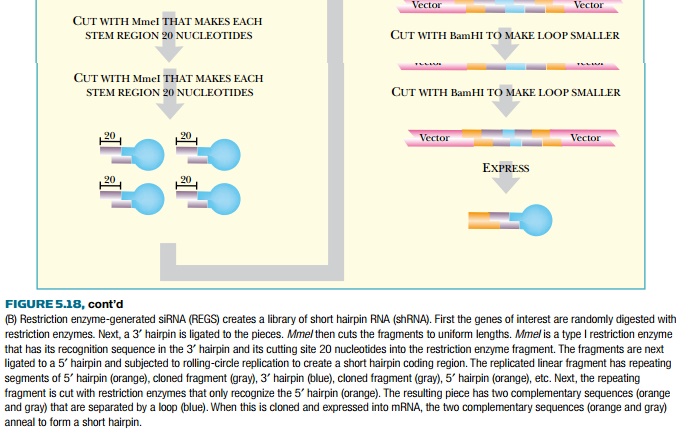
To make many siRNA copies of
these random constructs, they are converted into double-stranded loops. First,
a 5′ hairpin is ligated onto the
end cut by MmeI. A viral DNA
polymerase (from φ29) then converts the loop
into dsDNA. This polymerase can separate the strands in double-stranded regions
and so synthesizes a second strand for the entire circle. Since φ29 DNA polymerase displaces dsDNA, it continues
to circle around and around making a long concatemer for each loop, as in
rolling circle replication. The concatemer is digested with BglII and MlyI, to generate two complementary gene fragments connected by the
3′ loop. The 3′ loop is shortened by digestion with BamHI, and then the two fragments are
ligated together. The final product is cloned into a vector that will
transcribe the insert as a single strand of RNA. Finally, the two complementary
sequences base-pair to form a short hairpin that is processed into siRNA by
Dicer.
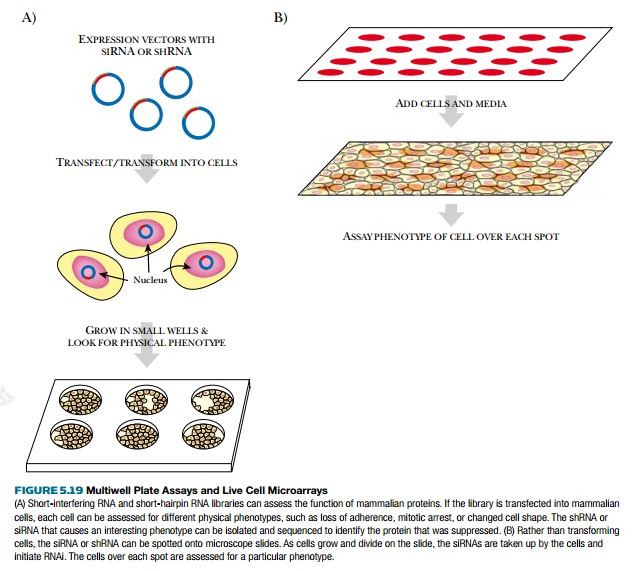
The RNAi libraries can be
analyzed either with multiwell plates or live
cell microarrays (Fig. 5.19). The multiwell plates use a barcode sequence on the shRNA in order
to identify which clone corresponds to which gene. A barcode or zipcode
sequence is approximately 20 nucleotides long and is unique to each clone. Each
clone can be identified by this sequence rather than sequencing the entire gene
insert. To analyze the library, it is transformed into a large number of cells
that are inoculated into the wells of a multiwell plate. A defined number of
cells go into each well, so
that only one siRNA is found in each well. The cells are then analyzed for any
noticeable symptoms. For those showing effects, the siRNA is identified by the
zipcode sequence. Another method for screening a siRNA or shRNA library is to
spot each clone onto slides in a microarray. Live cells are then added and take
up the siRNA or shRNA at these locations. The change in cell phenotype over a
spot corresponds to a particular siRNA.
If each protein in a genome
is studied for detectable phenotypes via RNAi, a phenotype catalogue can
be constructed. For example, many genes/proteins involved with cell membrane division will arrest cells with a
bridge structure during cell division. An unknown protein that causes the same
phenotype in an RNAi screen would potentially belong to the same family of
proteins. These proteins are said to have the same phenotypic signature. Databases have already been devised for C. elegans
(http://nematode.bio.nyu.edu:8001) and Drosophila
(http:// www.flyRNAi.org) where this type of information is compiled to share
with other scientists.
Related Topics