Chapter: Microbiology
Enzymology of DNA Replication
Enzymology of DNA Replication:
Replication of double stranded DNA molecule is a complex pro-cess involving a number of enzymes. For DNA replication to occur, the following events should take place
· Temporary separation of the two parental strands.
· Stabilization of the single stranded DNA molecule
· Initiation of daughter strand synthesis.
· Elongation of the daughter strands.
· Termination of the reaction
All the stages are individual enzymatic activities and do not func-tion independently and are contained in a discrete multiprotein struc-ture called the replisome. Enzymes that are able to synthesize new DNA strands on a template strand are called DNA polymerases.
The enzymes that polymerize nucleotides into a growing strand of DNA are called as polymerases. There are three known enzymes inE.coli
· DNA Polymerase I
· DNA Polymerase II
· DNA Polymerase III
In a simple model of DNA replication, according to the rule of complementarity, nucleotides will be synthesized on both the strands on the replication fork. During DNA replication polymerization pro-ceeds from 5’ to 3’ direction. Since both strands are running in oppo-site direction one new strand has to be replicated in the 5’ to 3’ direc-tion and the other in the 3’ to 5’ direction. However, all the known polymerases synthesise nucleotides only in the 5’ to 3’ direction. Evi-dence from autoradiography suggests that there are 2 types of replica-tion
· Continuous replication
· Discontinuous replication
The discontinuous form of replication takes place on the comple-mentary strand in short segments in a backward direction. These short segments are called as Okazaki fragments, named after R. Okazaki who first saw them. The length of Okazaki fragments in prokaryotes is 1500 nucleotides and 150 in eukaryotes. The strand that is synthesized continuously is called as leading strand. The discontinuous strand is called as lagging strand.
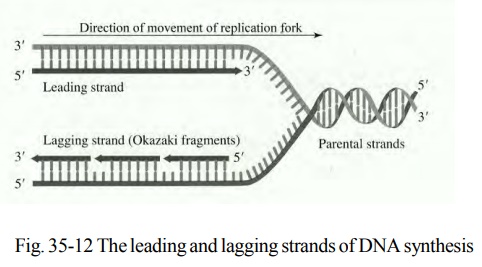
The list of major proteins necessary for DNA replication in E.coliis tabulated
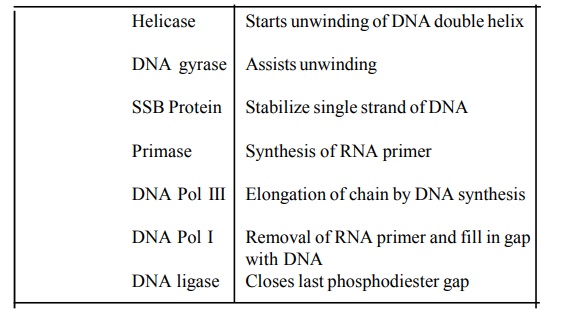
The combined effect of helicase and gyrase results in the forma-tion of a replication fork. (Figure 35-13
The helicase enzyme, (also called as unwinding protein and rep protein) recognizes and binds to the origin of replication and catalyses separation of the two DNA strands by breaking the hydrogen bonds between base pairs. DNA gyrase, a topoisomerase, assists unwinding of DNA strands by inducing supercoiling. The exposed single strands of DNA is stabilized and protected from hydrolytic cleavage of phosphodiester bonds. The SSB proteins (Single Stranded DNA Bind-ing protein) perform this protective role.
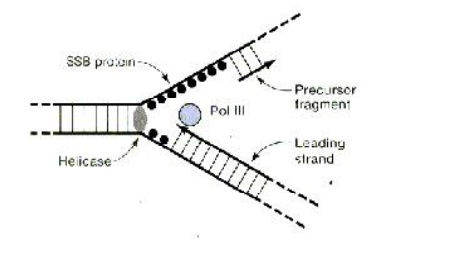
The various events of replication with essential enzymes and proteins
The separated polynucleotide strands are used as templates for the synthesis of complementary strands. The next step is the initiation of DNA synthesis. All of the known DNA polymerases can extend a deoxyribonucleotide chain from a free 3’ – OH end, but none can initiate synthesis. The DNA Polymerase III requires primer with a free3’-hydroxyl end. The primer is short stretch of RNA (4 to 10 nucle-otides) complementary to the DNA template.
RNA synthesis is catalysed by an enzyme called primase. The action of primase is required only once for the initiation of the leading strand of DNA where as each okazaki fragment must be initiated by the action of primase.
The DNA synthesis is catalysed by Polymerase III and can pro-ceed from 3’-hydroxyl group. The RNA primer is removed from theDNA by the 5’ - - > 3’ nuclease action of DNA Polymerase I and byRNAase H after the second synthesis is computed.
When the primer is removed, there will be a gap. DNA Poly-merase I is likely to be involved in filling the gap.
Both leading and lagging strand are extended in the 5’ - - > 3’ di-rection. The leading strand proceed in the direction of the advancing replication fork Synthesis of the lagging strand continues in the oppo-site direction until it meets the fragment previously synthesized.
Once the RNA has been removed and replaced, the adjacent Okazaki fragments must be linked together. The 3’-OH end of one fragment will be adjacent to the 5’ phosphate end of the previous frag-ment. The gap is filled by the enzyme DNA ligase by forming the final phosphodiester bond.
Termination of the replication process occurs when the two rep-lication forks meet in the circular E.coli chromosome.
Related Topics