Chapter: Biochemistry: The Three-Dimensional Structure of Proteins
Tertiary Structure of Proteins
Tertiary Structure of Proteins
The
tertiary structure of a protein is the three-dimensional arrangement of all the
atoms in the molecule. The conformations of the side chains and the positions
of any prosthetic groups are parts of the tertiary structure, as is the
arrangement of helical and pleated-sheet sections with respect to one another.
In a fibrous protein, the overall shape of which is a long rod, the secondary
structure also provides much of the information about the tertiary structure.
The helical backbone of the protein does not fold back on itself, and the only
important aspect of the tertiary structure that is not specified by the
secondary structure is the arrangement of the atoms of the side chains.
For a
globular protein, considerably more information is needed. It is neces-sary to
determine the way in which the helical and pleated-sheet sections fold back on each
other, in addition to the positions of the side-chain atoms and any prosthetic
groups. The interactions between the side chains play an important role in the
folding of proteins. The folding pattern frequently brings residues that are
separated in the amino acid sequence into proximity in the tertiary structure
of the native protein.
Forces Involved in Tertiary Structures
Many
types of forces and interactions play a role in holding a protein together in
its correct, native conformation. Some of these forces are covalent, but many
are not. The primary structure of a protein-the order of amino acids in the
polypeptide chain-depends on the formation of peptide bonds, which are
covalent. Higher-order levels of structure, such as the conformation of the
backbone (secondary structure) and the positions of all the atoms in the
protein (tertiary structure), depend on noncovalent interactions. If the
protein consists of several subunits, the interaction of the subunits also
depends on noncovalent interactions. Noncovalent stabilizing forces contribute
to the most stable structure for a given protein, the one with the lowest
energy.
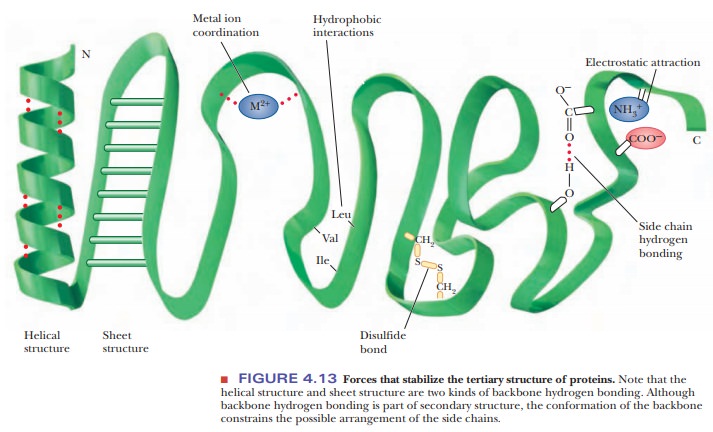
Several
types of hydrogen bonding occur in proteins. Backbone hydrogen bonding is a major determinant of secondary
structure; hydrogen bonds between the
side chains of amino acids are also possible in proteins. Nonpolar
resi-dues tend to cluster together in the interior of protein molecules as a
result of hydrophobic interactions. Electrostatic attraction between
oppositely charged groups, which frequently occurs on the surface of the
molecule, results in such groups being close to one another. Several side
chains can be complexed to a single
metal ion. (Metal ions also occur in some prosthetic groups.)
In
addition to these noncovalent interactions, disulfide
bonds form covalent links between the side chains of cysteines. When such
bonds form, they restrict the folding patterns available to polypeptide chains.
There are specialized labo-ratory methods for determining the number and positions
of disulfide links in a given protein. Information about the locations of
disulfide links can then be combined with knowledge of the primary structure to
give the complete covalentstructure of
the protein. Note the subtle difference here: The primary structureis the order
of amino acids, whereas the complete covalent structure also speci-fies the
positions of the disulfide bonds (Figure 4.13).
Not
every protein necessarily exhibits all possible structural features of the
kinds just described. For instance, there are no disulfide bridges in myoglobin
and hemoglobin, which are oxygen-storage and transport proteins and classic
examples of protein structure, but they both contain Fe(II) ions as part of a
prosthetic group. In contrast, the enzymes trypsin and chymotrypsin do not
contain complexed metal ions, but they do have disulfide bridges. Hydrogen
bonds, electrostatic interactions, and hydrophobic interactions occur in most
proteins.
The three-dimensional conformation of a protein is the result of the inter-play of all the stabilizing forces. It is known, for example, that proline does not fit into an α-helix and that its presence can cause a polypeptide chain to turn a corner, ending an α-helical segment. The presence of proline is not, however, a requirement for a turn in a polypeptide chain. Other residues are rou-tinely encountered at bends in polypeptide chains. The segments of proteins at bends in the polypeptide chain and in other portions of the protein that are not involved in helical or pleated-sheet structures are frequently referred to as “random” or “random coil.” In reality, the forces that stabilize each protein are responsible for its conformation.
How can the three-dimensional structure of a protein be determined?
The
experimental technique used to determine the tertiary structure of a protein is
X-ray crystallography. Perfect
crystals of some proteins can be grown under carefully controlled conditions.
In such a crystal, all the individual protein molecules have the same
three-dimensional conformation and the same orientation. Crystals of this
quality can be formed only from proteins of very high purity, and it is not
possible to obtain a structure if the protein cannot be crystallized.
When a
suitably pure crystal is exposed to a beam of X rays, a diffraction pat-tern is produced on a photographic plate (Figure
4.14a) or a radiation counter.The pattern is produced when the electrons in
each atom in the molecule scat-ter the X rays. The number of electrons in the
atom determines the intensity of its scattering of X rays; heavier atoms scatter
more effectively than lighter atoms. The scattered X rays from the individual
atoms can reinforce each other or cancel each other (set up constructive or
destructive interference), giving rise to the characteristic pattern for each
type of molecule. A series of diffrac-tion patterns taken from several angles
contains the information needed to determine the tertiary structure. The
information is extracted from the diffrac-tion patterns through a mathematical
analysis known as a Fourier series.
Many thousands of such calculations are required to determine the structure of
a pro-tein, and even though they are performed by computer, the process is a
fairly long one. Improving the calculation procedure is a subject of active
research.
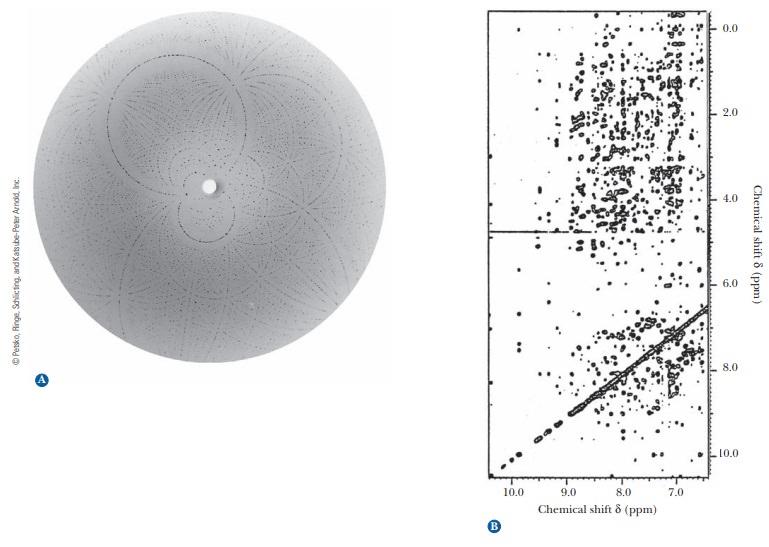
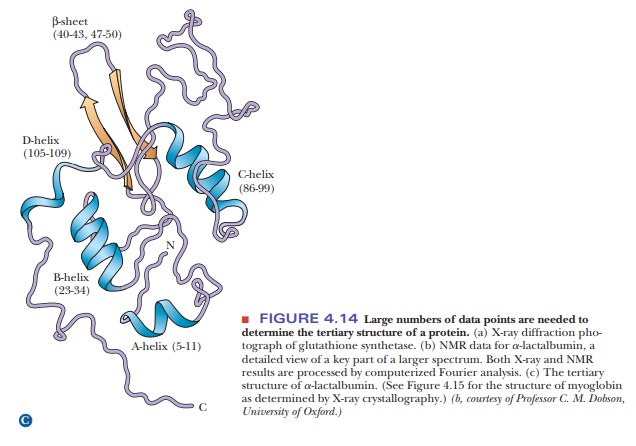
Another
technique that supplements the results of X-ray diffraction has come into wide
use in recent years. It is a form of nuclear
magnetic resonance(NMR) spectroscopy. In this particular application of
NMR, called2-D(twodimensional) NMR, large collections of data points
are subjected to computer analysis (Figure 4.14b). Like X-ray diffraction, this
method uses a Fourier series to analyze results. It is similar to X-ray
diffraction in other ways: It is a long pro-cess, and it requires considerable
amounts of computing power and milligram quantities of protein. One way in
which 2-D NMR differs from X-ray diffraction is that it uses protein samples in
aqueous solution rather than crystals. This environment is closer to that of
proteins in cells, and thus it is one of the main advantages of the method. The
NMR method most widely used in the deter-mination of protein structure
ultimately depends on the distances between hydrogen atoms, giving results
independent of those obtained by X-ray crystal-lography. The NMR method is
undergoing constant improvement and is being applied to larger proteins as
these improvements progress.
Myoglobin: An Example of Protein Structure
In many
ways, myoglobin is the classic example of a globular protein. We shall use it
here as a case study in tertiary structure. (We shall see the tertiary
structures of many other proteins in context when we discuss their roles in
biochemistry.) Myoglobin was the first protein for which the complete tertiary
structure (Figure 4.15) was determined by X-ray crystallography. The complete
myoglobin molecule consists of a single polypeptide chain of 153 amino acid
residues and includes a prosthetic group, the heme group, which also occurs in hemoglobin. The myoglobin molecule
(including the heme group) has a compact structure, with the interior atoms
very close to each other. This structure provides examples of many of the
forces responsible for the three-dimensional shapes of proteins.
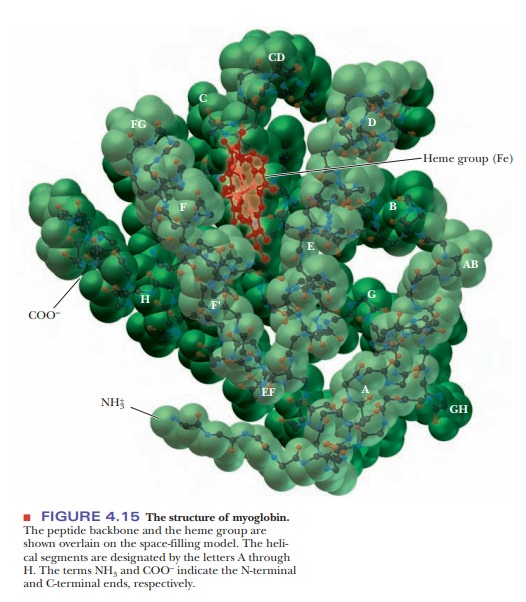
Myoglobin has eight α-helical regions and no β-pleated sheet regions. Approximately 75% of the residues in myoglobin are found in these helical regions, which are designated by the letters A through H. Hydrogen bonding in the polypeptide backbone stabilizes the α-helical regions; amino acid side chains are also involved in hydrogen bonds. The polar residues are on the exterior of the molecule.
The interior of the protein contains almost exclu-sively
nonpolar amino acid residues. Two polar histidine residues are found in the
interior; they are involved in interactions with the heme group and bound
oxygen, and thus play an important role in the function of the molecule. The
planar heme group fits into a hydrophobic pocket in the protein portion of the
molecule and is held in position by hydrophobic attractions between heme’s
porphyrin ring and the nonpolar side chains of the protein. The presence of the
heme group drastically affects the conformation of the polypeptide: The apoprotein
(the polypeptide chain alone, without the prosthetic heme group) is not as
tightly folded as the complete molecule.
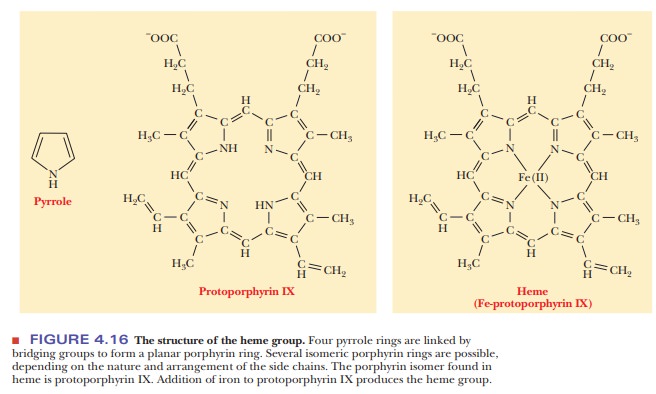
The heme
group consists of a metal ion, Fe(II), and an organic part, pro-toporphyrin IX
(Figure 4.16). (The notation Fe(II) is preferred to Fe2+ when
metal ions occur in complexes.) The porphyrin part consists of four
five-membered rings based on the pyrrole structure; these four rings are linked
by bridging methine (-CH==) groups to form a square planar structure. The
Fe(II) ion has six coordination sites, and it forms six metal–ion complexation
bonds. Four of the six sites are occupied by the nitrogen atoms of the four
pyr-role-type rings of the porphyrin to give the complete heme group. The
pres-ence of the heme group is required for myoglobin to bind oxygen.
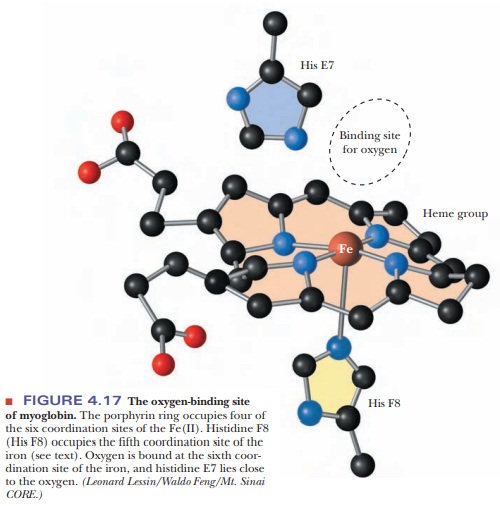
The
fifth coordination site of the Fe(II) ion is occupied by one of the nitrogen
atoms of the imidazole side chain of histidine residue F8 (the eighth residue
in helical segment F). This histidine residue is one of the two in the interior
of the molecule. The oxygen is bound at the sixth coordination site of the
iron. The fifth and sixth coordination sites lie perpendicular to, and on
opposite sides of, the plane of the porphyrin ring. The other histidine residue
in the interior of the molecule, residue E7 (the seventh residue in helical
seg-ment E), lies on the same side of the heme group as the bound oxygen
(Figure 4.17). This second histidine is not bound to the iron, or to any part
of the heme group, but it acts as a gate that opens and closes as oxygen enters
the hydro-phobic pocket to bind to the heme. The E7 histidine sterically
inhibits oxygen from binding perpendicularly to the heme plane, with
biologically important ramifications.
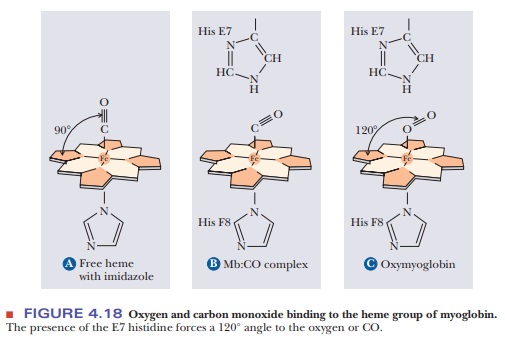
Why does oxygen have imperfect binding to the heme group?
At
first, it would seem counterintuitive that oxygen would bind imperfectly to the
heme group. After all, the job of both myoglobin and hemoglobin is to bind to
oxygen. Wouldn’t it make sense that oxygen should bind strongly? The answer
lies in the fact that more than one molecule can bind to heme. Besides oxygen,
carbon monoxide also binds to heme. The affinity of free heme for carbon
monoxide (CO) is 25,000 times greater than its affinity for oxygen. When carbon
monoxide is forced to bind at an angle in myoglobin because of the steric block
by His E7, its advantage over oxygen drops by two orders of magnitude (Figure
4.18). This guards against the possibility that traces of CO produced during
metabolism would occupy all the oxygen-binding sites on the hemes.
Nevertheless, CO is a potent poison in larger quantities because of its effect
both on oxygen binding to hemoglobin and on the final step of the electron
transport chain. It is also important to remember that although our metabolism
requires that hemoglobin and myoglobin bind oxygen, it would be equally
disastrous if the heme never let the oxygen go. Thus having binding be too
perfect would defeat the purpose of having the oxygen-carrying proteins.
In the
absence of the protein, the iron of the heme group can be oxidized to Fe(III);
the oxidized heme will not bind oxygen. Thus, the combination of both heme and
protein is needed to bind O2 for oxygen storage.
Denaturation and Refolding
The noncovalent interactions that maintain the three-dimensional structure of a protein are weak, and it is not surprising that they can be disrupted easily.
The
unfolding of a protein (i.e., disruption of the tertiary structure) is called denaturation. Reduction of disulfide
bonds leads to even moreextensive unraveling of the tertiary structure.
Denaturation and reduction of disulfide bonds are frequently combined when
complete disruption of the tertiary structure of proteins is desired. Under
proper experimental conditions, the disrupted structure can then be completely
recovered. This process of denaturation and refolding is a dramatic
demonstration of the relationship between the primary structure of the protein
and the forces that determine the tertiary structure. For many proteins,
various other factors are needed for complete refolding, but the important
point is that the primary structure determines the tertiary structure.
Proteins
can be denatured in several ways. One is heat.
An increase in tem-perature favors vibrations within the molecule, and the
energy of these vibra-tions can become great enough to disrupt the tertiary
structure. At either high or low extremes
of pH, at least some of the charges on the protein are missing, and so the
electrostatic interactions that would normally stabilize the native, active
form of the protein are drastically reduced. This leads to denaturation.
The
binding of detergents, such as sodium
dodecyl sulfate (SDS), also dena-tures proteins. Detergents tend to disrupt
hydrophobic interactions. If a deter-gent is charged, it can also disrupt
electrostatic interactions within the protein. Other reagents, such as urea and guanidine hydrochloride, form hydrogen bonds with the protein that
are stronger than those within the protein itself. These two reagents can also
disrupt hydrophobic interactions in much the same way as detergents (Figure
4.19).
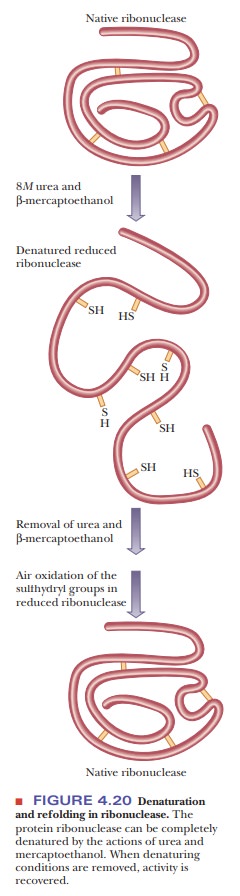
b-Mercaptoethanol (HS-CH2-CH2-OH) is
frequently used to reduce disul-fide bridges to two sulfhydryl groups. Urea is
usually added to the reaction mixture to facilitate unfolding of the protein
and to increase the accessibility of the disulfides to the reducing agent. If
experimental conditions are properly chosen, the native conformation of the
protein can be recovered when both mercaptoethanol and urea are removed (Figure
4.20). Experiments of this type provide some of the strongest evidence that the
amino acid sequence of the protein contains all the information required to
produce the complete three-dimensional structure. Protein researchers are
pursuing with some interest the conditions under which a protein can be
denatured-including reduction of disulfides-and its native conformation later recovered.
Summary
Tertiary structure is the complete
three-dimensional arrangement of all the atoms in a protein.
The tertiary structure of proteins is
maintained by different types of cova-lent and noncovalent interactions.
Hydrogen bonding occurs between atoms on the
peptide backbone as well as atoms in the side chains.
Electrostatic attractions between positively
charged side chains and nega-tively charged side chains are also important.
The tertiary structure of proteins is
determined by the techniques of X-ray diffraction and nuclear magnetic
resonance.
Myoglobin, the first protein to have its
tertiary structure determined, is a globular protein for oxygen storage.
Myoglobin is a single polypeptide chain with 153
amino acids, 8 α-helical regions, and a prosthetic group called heme.
The heme has a coordinated iron ion at the
center that binds to oxygen.
Proteins can be denatured by heat, pH, and
chemicals. Denaturation causes the protein to lose its native tertiary
structure.
Some
types of denaturation can be reversed, while others are permanent.
Related Topics