Chapter: Biochemistry: Nucleic Acid Biotechnology
Sequencing DNA
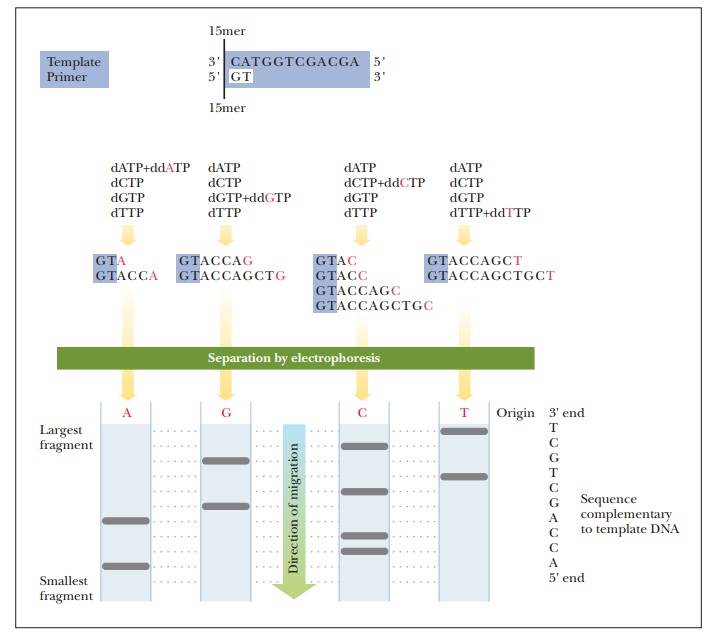
Sequencing DNA
We have already seen that the primary structure of a protein
determines its secondary and tertiary structures. The same is true of nucleic
acids; the nature and order of monomer units determine the properties of the
whole molecule. Base pairing in both RNA and DNA depends on a series of
complementary bases, whether these bases are on different polynucleotide
strands, as in DNA, or on the same strand, as is frequently the case in RNA.
Sequencing of nucleic acids is now fairly routine, and this relative ease would
have amazed the scientists of the 1950s and 1960s.
The method devised by Sanger and Coulson for determining the base sequences of nucleic acids depends on selective interruption of oligonucle-otide synthesis. A single-stranded DNA fragment whose sequence is to be determined is used as a template for the synthesis of a complementary strand. The new strand grows from the 5' end to the 3' end. This unique direction of growth is true for all nucleic acid synthesis. The synthesis is inter-rupted at every possible site in a population of molecules. The interruption of synthesis depends on the presence of 2',3'-dideoxyribonucleoside triphos-phates (ddNTPs).
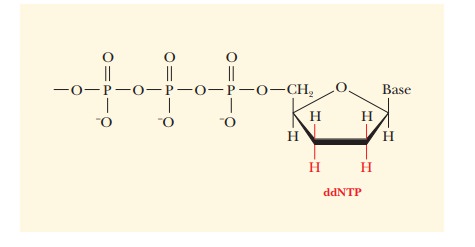
The 3'-hydroxyl group of deoxyribonucleoside triphosphates (the
usual monomer unit for DNA synthesis) has been replaced by a hydrogen.
How do dideoxy nucleotides allow us to sequence DNA?
These ddNTPs can be incorporated in a growing DNA chain, but they
lack a 3'-hydroxyl group to form a bond to another nucleoside triphosphate. The
incorporation of a ddNTP into the growing chain causes termination at that
point. The presence of small amounts of ddNTPs in a replicating mixture causes
random termination of chain growth.
The DNA to be sequenced is mixed with a short oligonucleotide that
serves as a primer for synthesis of the complementary strand. The primer is
hydrogen-bonded toward the 3' end of the DNA to be sequenced. The DNA with
primer is divided into four separate reaction mixtures. Each reaction mixture
contains all four deoxyribonucleoside triphosphates (dNTPs), one of which is
labeled to allow the newly synthesized fragments to be visualized by
autoradiography or by fluorescence. In addition, each of the reaction mixtures
contains one of the four ddNTPs. Synthesis of the chain is allowed to proceed
in each of the four reaction mixtures. In each mixture, chain termina-tion
occurs at all possible sites for that nucleotide.
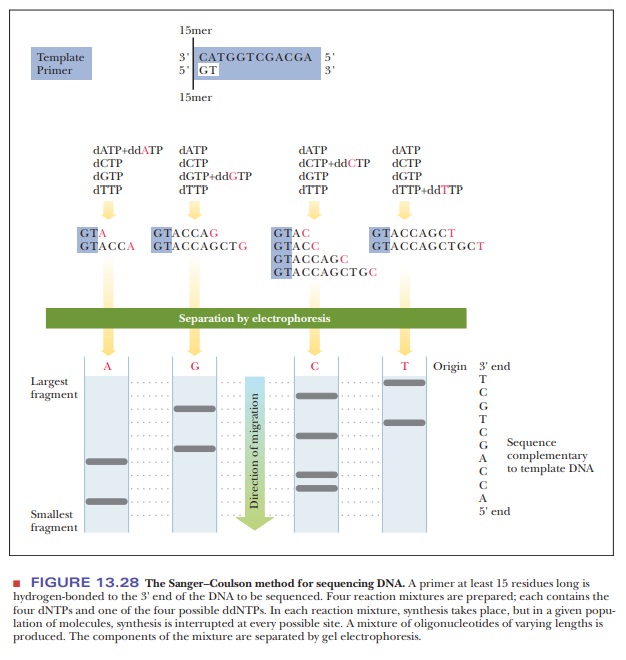
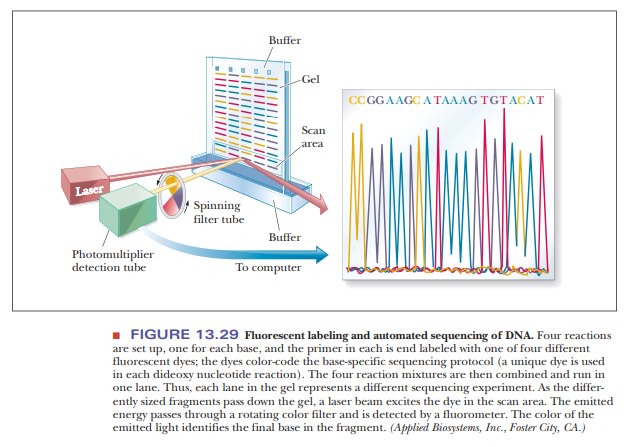
When gel electrophoresis is performed on each reaction mixture, a band corresponding to each position of chain termination appears. The sequence of the newly formed strand, which is complementary to that of the template DNA, can be “read” directly from the sequencing gel (Figure 13.28). A variation on this method is to use a single reaction mixture with a different fluorescent label on each of the four ddNTPs. Each fluorescent label can be detected by its characteristic spectrum, requiring only a single gel electrophoresis experiment.
The use of fluorescent labels makes it possible to automate DNA
sequencing, with the whole process under computer control. Commercial kits are
available for these sequencing methods (Figure 13.29).
When RNA is to be sequenced, the method of choice is not to analyze
the RNA itself but to use the methods of DNA sequencing on a DNA complemen-tary
(cDNA) to the RNA in question. The cDNA, in turn, is generated by using the
enzyme reverse transcriptase, which catalyzes the synthesis of DNA from an RNA
template.
Summary
DNA can be sequenced by using several techniques. One of the most
com-mon is called the chain termination method.
Dideoxy nucleotides are used to terminate DNA synthesis. Multiple
reac-tions are run with a different dideoxy nucleotide in each reaction mix.
The reactions produce a series of DNA fragments of different length
that can be run on a gel and the sequence determined by tracking the different
length fragments in the lanes with the four different dideoxy nucleotides.
Related Topics