Chapter: Biochemistry: Nucleic Acid Biotechnology
Genomics and Proteomics
Genomics and Proteomics
With more and more full DNA sequences becoming available, it is
tempting to compare those sequences to see whether patterns emerge from genes
that encode proteins with similar functions. The amount of data makes use of a
computer essential for the process. Databases on genome and protein sequences
are so extensive as to require information technology at its best to solve
problems. Knowing the full DNA sequence of the human genome, for example,
allows us to address the causes of disease in a way that was not possible until
now. That prospect was one of the main incentives for undertaking the Human
Genome Project. The website of the National Human Genome Research Institute,
which is a part of the National Institutes of Health (NIH) has useful information;
the URL for this site is http :// www .genome.gov.
A number of genomes are available online, along with software for
sequence comparisons. An example is the material available from the Sanger
Institute (http :// www .sanger.ac.uk). In November 2003, the researchers at
this institute announced that they had sequenced 2 billion bases from the DNA
of several organisms (human, mouse, zebrafish, yeasts, and the roundworm Caenorhabditiselegans, among others). If
this amount of DNA were the size of a spiral staircase,it would reach from the
Earth to the Moon.
A question implicit in the determination of the genome of any
organism is that of assignment of sequences to the chromosome in which they
belong. This is a challenging task, and only suitable computer algorithms make
it possible. Once this has been achieved, one can compare genomes to see what
changes have occurred in the DNA of complex organisms compared with those of
sim-pler ones.
Beyond this application, challenging though it may be, lies the application
to medicine, which is leading to a number of surprises. Two closely related
genes (BRCA1 and BRCA2) involved in the development of breast cancer inter-act with
other genes and proteins, and this is a topic of feverish research. The
connection between these genes and a number of seemingly unrelated cancers is
only starting to be unraveled. Clearly, there is need to determine not only the
genetic blueprint but the manner in which an organism puts it into action.
The proteome is the
protein version of the genome. In all organisms for which sequence information
is available, proteomics (the study
of interac-tions among all the proteins in a cell) is assuming an important
place in the life sciences. If the genome is the script, the proteome puts the
play on stage. The genetically determined amino acid sequence of proteins
determines their structure and how they interact with each other. Those
interactions determine how they behave in a living organism. The potential
medical applications of the human proteome are apparent, but these have not yet
been realized. Proteomic information does exist, for eukaryotes such as yeast
and the fruit fly Drosophila
melanogaster, and the methods that have been developed for thoseexperiments
will be useful in unraveling the human proteome.
The Power of Microarrays—Robotic Technology Meets Biochemistry
Thousands of genes and their products (i.e., RNA and proteins) in a
given living organism function in a complicated and harmonious way.
Unfortunately, traditional methods in molecular biology have always focused on
analyzing one gene per experiment. In the past several years, a new technology,
called DNAmicroarray (DNA chip or gene chip), has attracted tremendous interest amongmolecular
biologists. Microarrays allow for the analysis of an entire genome in one
experiment and are used to study gene expression, the transcription rates of
the genome in vivo. The genes that are being transcribed at any particular time
are known as the transcriptome. The
principle behind the microarray is the placement of specific nucleotide
sequences in an ordered array, which then base-pair with complementary
sequences of DNA or RNA that have been labeled with fluorescent markers of
different colors. The locations where binding occurred and the colors observed
are then used to quantify the amount of DNA or RNA bound. Microarray chips are
manufactured by high-speed robotics, which can put thousands of samples on a
glass slide with an area of about 1 cm2. The
diameter of an individual sample might be 200 μm or less. Several different methods are used
for implanting the DNA to be studied on the chip, and many companies make
microarray chips.
How do microarrays work?
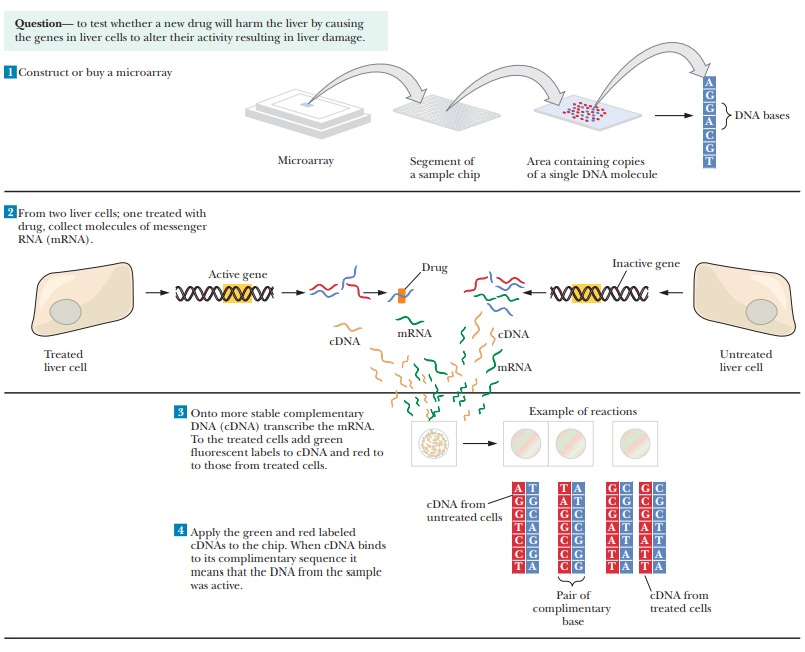
Figure 13.30 shows an example of how microarrays could be used to
determine whether a potential new drug would be harmful to liver cells. In Step
1, a microarray is purchased or constructed that has single-stranded DNA
representing thousands of different genes, each applied at a specific spot on
the microarray chip. In Step 2, different populations of liver cells are
collected, one treated with the potential drug and the other untreated. The
mRNA being transcribed in these cells is then collected. In Step 3, the mRNA is
converted to cDNA. Green fluorescent labels are added to the cDNA from the
untreated cells, and red fluorescent labels are added to the cDNA from the
treated cells. In Step 4, the labeled cDNAs are added to the chip. The cDNAs
bind to the chip if they find their complementary sequences in the
single-stranded DNAs loaded on the chip. The expanded portion of Step 4 shows
what is happening at the molecular level. The black sequences represent the DNA
bound to the chip. Green sequences represent the cDNA from the untreated cells
that bind to their target sequences. The red sequences are cDNA from treated
cells. Some of the DNA sequences on the chip bind nothing. Some bind only the
red sequences while others bind only the green ones. Some sequences on the chip
bind both.
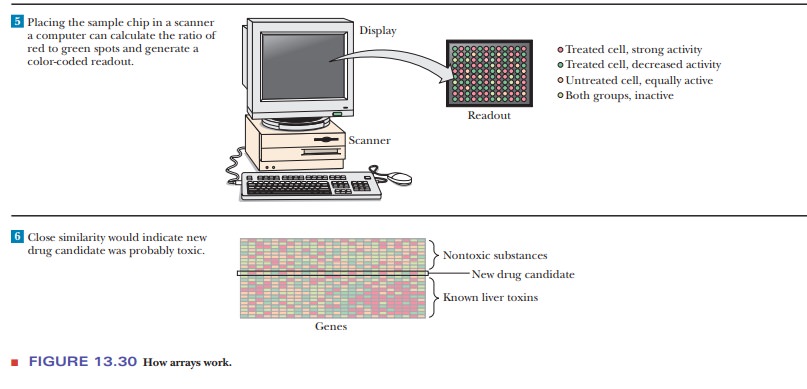
In Step 5, the chip is scanned and a computer analyzes the
fluorescence. The results appear as a series of colored dots. A red dot
indicates a DNA sequence on the chip that bound to the cDNA from the treated
cells. This indicates an mRNA that was being expressed in treated cells. A
green dot indicates RNA produced in untreated but not treated cells. A yellow
dot would indicate a mRNA that was produced equally well in treated or
untreated cells. Black spaces indicate DNA sequences on the chip for which no
mRNA was produced in either situation. To answer the question about whether the
potential drug is toxic to liver cells, the results from the microarray would
be compared to controls run with liver cells and drugs known to be toxic versus
those known to be nontoxic, as shown in Step 6.
Figure 13.31 shows the results of a study designed to scan cells
from cancer patients and correlate microarray patterns with prognoses. The four
different patterns are compared to the percentage of patients who developed
metastases. Information like this could be critical to treatment of cancer.
Doctors often have to choose between different strategies. If they had access
to data like this from their patients, they would be able to predict the
likelihood of the patient’s developing more serious forms of cancer, and
thereby able to choose a more appropriate treatment.
Protein Arrays
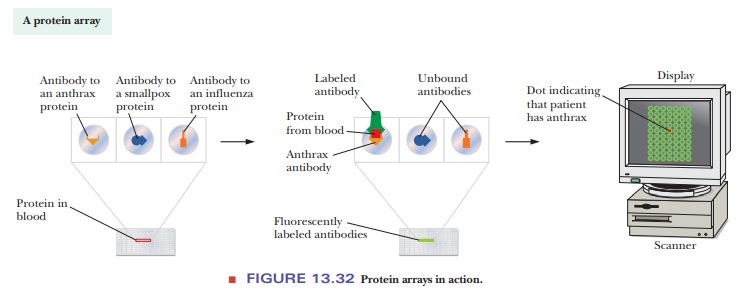
Another type of microchip uses bound proteins instead of DNA. These
protein arrays are based on interactions between proteins and antibodies . For
example, antibodies to known diseases can be bound to the microarray. A sample
of a patient’s blood can then be put on the microarray. If the patient has a
particular disease, proteins specific to that disease bind to the appropriate antibodies.
Fluorescently labeled antibodies are then added and the microarray scanned. The
results look similar to the DNA microarrays discussed previously. Figure 13.32
shows how this would work to identify that a patient had anthrax. This
technique is growing in popularity and power, but is limited by whether
purified antibodies have been created for a particular disease.
Summary
As more DNA sequences become available, it becomes possible to
com-pare those sequences. Of particular interest is any pattern that may emerge
from genes that encode proteins with similar functions.
Important medical applications are emerging, and new
methods are mak- ing it possible to analyze large quantities of data. Complete
protein–protein interaction maps are now available for eukaryotes.
The proteome is the protein version of the genome. It refers to all
the proteins being expressed in a cell. The study of proteomics is becoming
very important in the life sciences.
A very powerful technique in vogue these days is the use of DNA or
protein microchips. With these chips, thousands of samples of DNA or proteins
can be applied and then checked for binding of biological samples.
The binding is visualized by using fluorescently labeled molecules
and scanning the chip with a computer. The pattern of fluorescent labels then
indicates which mRNA or proteins are being expressed in the samples.
Related Topics