Chapter: Biochemistry: Nucleic Acid Biotechnology
DNA Libraries
DNA Libraries
Because methods exist for selecting regions of DNA from the genome
of an organism of interest and cloning those regions in suitable vectors, a
question that immediately comes to mind is whether we can take all the DNA of an organism (the total
genome) and clone it in chunks of reasonable size. The answer is that we can do
this, and the result is a DNA library.
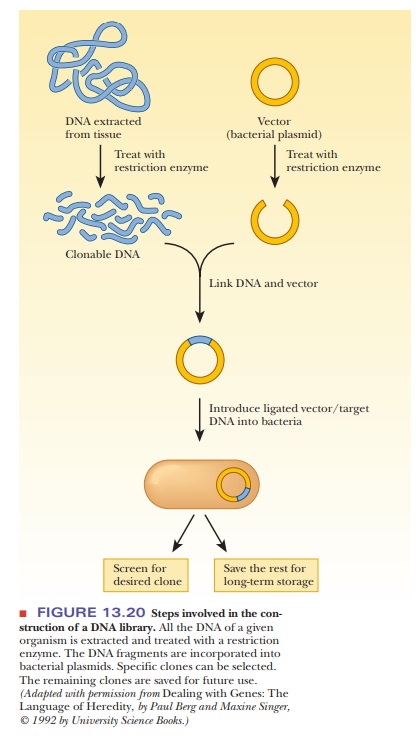
Let’s say that we want to construct a library of the human genome.
A diploid human cell (a cell that has a set of chromosomes from both parents)
has six billion base pairs. If we consider that 20,000 base pairs is a
reasonable size for a cloned insert, we need a minimum of 300,000 different
recombinant DNAs. It is quite a feat to achieve this number of different
recombinants, and, in prac-tice, we would need several times this minimum to
ensure full representation and to account for the vector molecules that do not
acquire an insert. For the purposes of this discussion, we shall use a
bacterial plasmid as the vector to construct a suitable number of recombinant DNA
molecules by the methods described (Figure 13.20).
The next step is to separate the individual members of the population of plasmid DNA molecules by cloning them. The group of clones that has acquired a plasmid with an insert constitutes the recombinant library. The whole library can be stored for future use, or a single clone can be selected for further study. The process of constructing a DNA library can be quite labori-ous, leading many researchers to obtain previously constructed libraries from other laboratories or from commercial sources. Some journals require that libraries and individual clones that have been discussed in articles they publish be freely available to other laboratories.
How do we find the piece of DNA we want in a library?
Finding an Individual Clone in a DNA Library
Imagine that, after a DNA library has been constructed, someone
wants to find a single clone (for example, one that contains a gene responsible
for an inherited disease) out of the hundreds of thousands, or possibly
millions, in the library. This degree of selectivity requires specialized
techniques. One of the most useful of these techniques depends on separating
and annealing complementary strands. An imprint is taken of the petri dish on
which the bacterial colonies (or phage plaques) have grown. A nitrocellulose
disc is placed on the dish and then removed. Some of each colony or plaque is
transferred to the disc, and theposition
of each is the same as it was on the dish. The rest of the original
colonies orplaques remain on the dish and can be stored for future use (Figure
13.21).
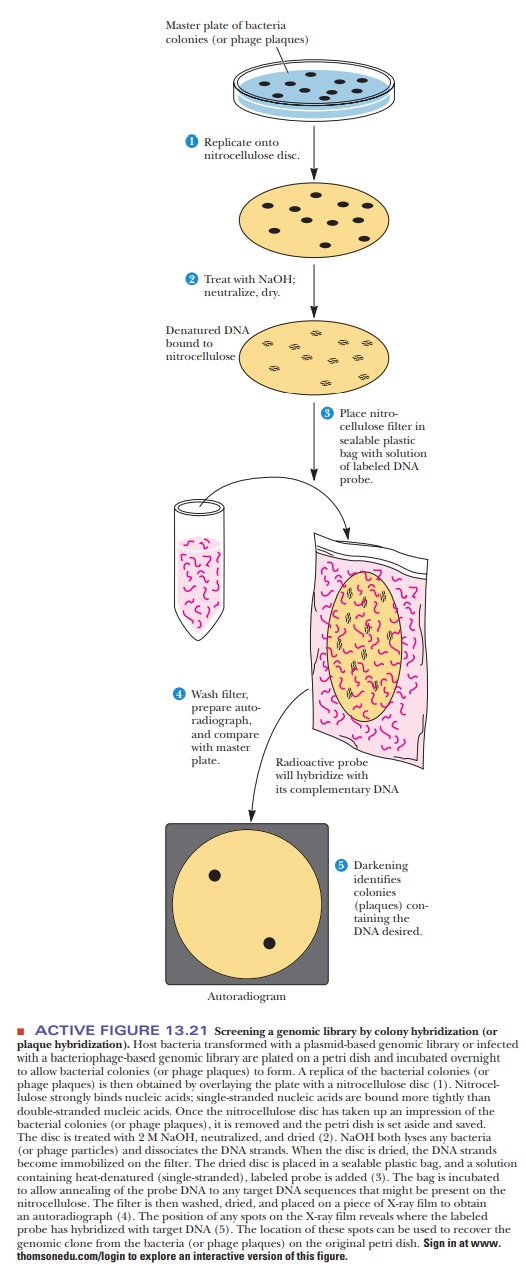
The nitrocellulose disc is treated with a denaturing agent to unwind all the DNA on it. [The DNA has become accessible by disruption (lysis) of the bacte-rial cells or phage.] After it is denatured, the DNA is permanently fixed to the disc by treatment with heat or ultraviolet light. The next step is to expose the disc to a solution that contains a single-stranded DNA (or RNA) probe that has a sequence complementary to one of the strands in the clone of interest (Figure 13.21).
The probe anneals to the DNA of interest and only
to that DNA. Any excess solution is washed off the nitrocellulose disc. In the
case of radioactive probes, the disc is placed in contact with X-ray film. Only
the spots on the disc in which some of the probe has annealed to the DNA
already there are radioactive, and only those spots expose the X-ray film. Because
the original petri dish has been saved, the desired clone can be picked off the
plate and allowed to reproduce.
If the nucleotide sequence of the desired DNA segment is not known
and no probe is available, a complication arises. If the gene of interest
directs the synthesis of a given protein, one chooses a vector that allows
cloned genes to be transcribed and translated. If the presence of the desired
protein can be detected by its function, that serves as the basis for detecting
it. Alternatively, labeled antibodies can be used as a basis for protein
detection.
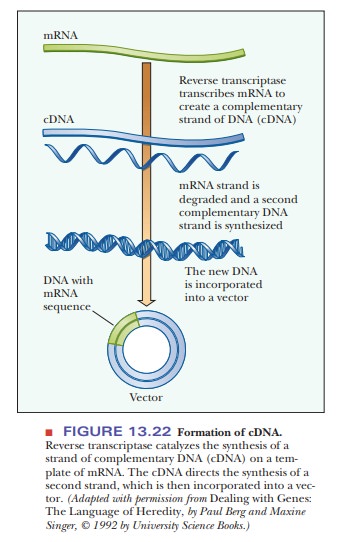
RNA libraries are not constructed and cloned as such. Rather, the
RNA of interest (usually mRNA) is used as the template for synthesis of
complemen-tary DNA (cDNA) in a reaction catalyzed by reverse transcriptase. The
cDNA is incorporated into a vector (Figure 13.22). Ligating the cDNA to a
vector requires the use of a synthetic linker. From this point on, the process
of pro-ducing a cDNA library is
virtually identical to that for constructing a genomic DNA library. For a given
organism, a genomic DNA library is the same no mat-ter what the tissue source,
and the DNA represents both expressed and unex-pressed DNA. In contrast, a cDNA
library is different depending on the tissue used and the expression profile of
the cells.
Summary
■ A DNA library is a collection of clones of an entire genome.
■ The genome is digested with restriction enzymes and the pieces are
cloned into vectors and transformed into cell lines.
■ Specific radioactive probes to a sequence of interest are reacted
to filters that have copies of the bacterial colonies in the library. The
probe bindsto the sequence of interest, and the colony’s location can be seen
viaautoradiography.
■ A cDNA library is constructed by using reverse transcriptase to
make DNA from the mRNA in a cell. This cDNA is then used to construct a
library similar to a genomic DNA library.
Related Topics