Chapter: Biochemistry: Nucleic Acid Biotechnology
RNA Interference - The Newest Way to Study Genes
RNA
Interference - The Newest Way to Study Genes
RNA interference (RNAi) was first discovered in a nematode worm (Caenorhabditis elegans). Double-stranded
RNA (dsRNA) was found to cause gene silencing in a sequence-specific way.
Researchers had long thought that RNA would be the perfect way to
control gene expression, because the right sequence of RNA should bind to DNA
and interfere with its transcription. While checking on the efficiency of
antisense RNA as a suppressor of gene expression, researchers discovered that
dsRNA was more than 10 times as effective at shutting off transcription of a
gene. In addition, suppression by RNAi was shown to be transmissible to other
cells of the organism and to the progeny in C.
elegans. RNAi has since been found as a natu-ral phenomenon in plants as a
way of targeting viral RNAs for destruction. It has also been seen in Drosophila and, most recently, in
mammals. It is now believed that RNAi is a natural regulatory mechanism for
controlling gene expression. It may also be a pro-tection mechanism against
oncogenes that produce too much of a harmful product.
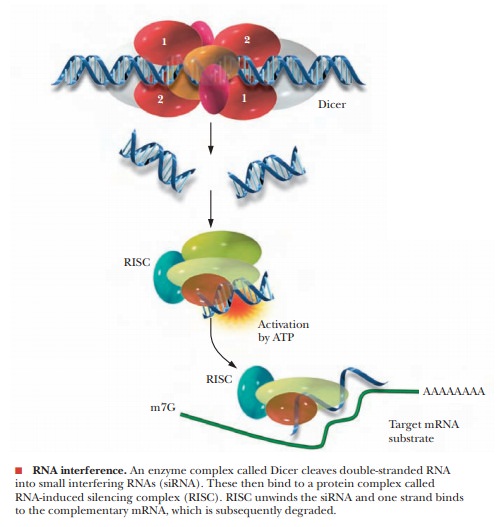
The process starts with an enzyme that is a member of the RNase III class, called Dicer. This enzyme binds to dsRNA and cleaves it into small interfering RNA (siRNA) of between 22 and 25 nucleotides, as shown in the figure.
The siRNAs then bind to a protein complex called RNA-induced silencing complex (RISC). The
siRNA–RISC complex then binds to target mRNAthat has the same sequence as part
of the siRNA and degrades it. In this way, the sequence of the dsRNA controls
the degrada-tion of an mRNA target. We are still learning how this may have
arisen naturally. Perhaps an endogenous RNA-dependent RNA polymerase senses
that too much of a particular mRNA is being made. It could then create the
opposite strand, thereby forming the dsRNA. This would then trigger the
eventual destruction of the mRNA. The same logic could apply to a defense
mechanism against viruses.
Regardless of RNAi’s natural purpose, it has become the
fastest-growing new field in molecular biology. Many companies have sprung up
overnight that produce RNAi kits and dsRNAs to use to initiate reactions. The
technique is quickly becoming the newest way to knock out specific genes to see
what then happens to the organism. All it takes is a knowledge of the gene
sequence, and the correct dsRNA can then be used to produce siRNAs to shut off
the gene. Researchers using RNAi have been better able to map the thousands of
genes in certain organisms, such as C.
elegans. As an example of the power of the technique, researchers just
mapped the gene for an enzyme involved in the metabolism of vitamin K. This
enzyme, vitamin K epoxide reduc-tase
(VKOR), is the target of a widely prescribed anticoagulant.The enzyme has
been known for 40 years, but attempts to purify it and locate it on the human
chromosome had met with no success. Using information from the Human Genome
Project, researchers were able to map human chromosomes and to come up with 13
genes that had characteristics that could mean that they were the VKOR gene.
Using RNAi, the correct one was iso-lated in a few weeks, a major breakthrough
for researchers that had been studying this enzyme for decades.
Medical researchers are very hopeful about the possibilities for
this technique. Given the specificity of RNAi, mutant alleles that are the
basis of diseases could be knocked out with the right sequence of dsRNA given
as a trigger for the Dicer enzyme to pro-cess. For example, if one allele is
mutated but the other is normal, RNAi could knock out the mutated one while not
affecting the normal one.
Related Topics