Chapter: Biochemistry: Nucleic Acid Biotechnology
Restriction Endonucleases
Restriction Endonucleases
Many
enzymes act on nucleic acids. A group of specific enzymes acts in concert to
ensure the faithful replication of DNA, and another group directs the
transcription of the base sequence of DNA into that of RNA. Other enzymes,
called nucleases, catalyze the
hydrolysis of the phosphodiester backbones of nucleic acids. Some nucleases are
specific for DNA; others are specific for RNA. Cleavage from the ends of the
molecule (by exonucleases) is known,
as is cleavage in the middle of the chain (by endonucleases). Some enzymes are specific for single-stranded
nucleic acids, and others cleave double-stranded ones. One group of nucleases, restriction endonucleases, has played a
crucial role in the development of recombinant DNA technology.
Restriction
enzymes were discovered in the course of genetic investigations of bacteria and
bacteriophages (phages for short; from the Greek phagein,“to eat”), the viruses that infect bacteria. The
researchers noted that bacte-riophages that grew well in one strain of the
bacterial species they infected frequently grew poorly (had restricted growth) in another strain of
the same species. Further work showed that this phenomenon arises from a subtle
differ-ence between the phage DNA and the DNA of the strain of bacteria in
which phage growth is restricted. This difference is the presence of methylated
bases at certain sequence-specific sites in the host DNA and not in the viral
DNA.
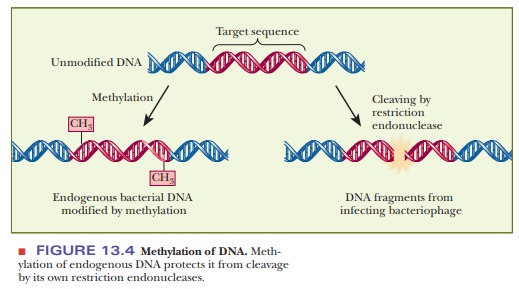
The
growth-restricting host cells contain cleavage enzymes, the restriction
endonucleases, that produce double-chain breaks at the unmethylated specific
sequences in phage DNA; the cells’ own corresponding DNA sequences, in which
methylated bases occur, are not attacked, as shown in Figure 13.4. These
cleavage enzymes consequently degrade DNA from any source but the host cell. The most immediate consequence is a slowing of
the growth of the phage in that bacterial strain, but the important thing for
our discussion is that DNA from any source can be cleaved by such an enzyme if
it contains the target sequence. More than 800 restriction endonucleases have
been discovered in a variety of bacterial species. More than 100 specific
sequences are recognized by one or more of these enzymes. Table 13.1 shows
several target sequences.

Many Restriction Endonucleases Produce “Sticky Ends”
Each restriction endonuclease hydrolyzes only a specific bond of a specific sequence in DNA. The sequences recognized by restriction endonucleases- their sites of action-read the same from left to right as they do from right to left (on the complementary strand). The term for such a sequence is a palindrome. (“Able was I ere I saw Elba” and “Madam, I’m Adam” are well-known linguistic palindromes). A typical restriction endonuclease called EcoRI is isolated from E. coli (each restriction endonuclease is designated by an abbreviation of thename of the organism in which it occurs). The EcoRI site in DNA is 5'-GAATTC-3', where the base sequence on the other strand is 3'-CTTAAG- 5'. The sequence from left to right on one strand is the same as the sequence from right to left on the other strand. The phosphodiester bond between G and A is the one hydrolyzed. This same break is made on both strands of the DNA. There are four nucleotide residues-two adenines and two thymines in each strand- between the two breaks on opposite strands, leaving sticky ends, which can still be joined by hydrogen bonding between the complementary bases. With the ends held in place by the hydrogen bonds, the two breaks can then be resealed covalently by the action of DNA ligases (Figure 13.5). If no ligase is present, the ends can remain separated, and the hydrogen bonding at the sticky ends holds the molecule together until gentle warming or vigorous stirring effects a separation. Some enzymes, such as HaeIII, cut in a way that leave a blunt end.
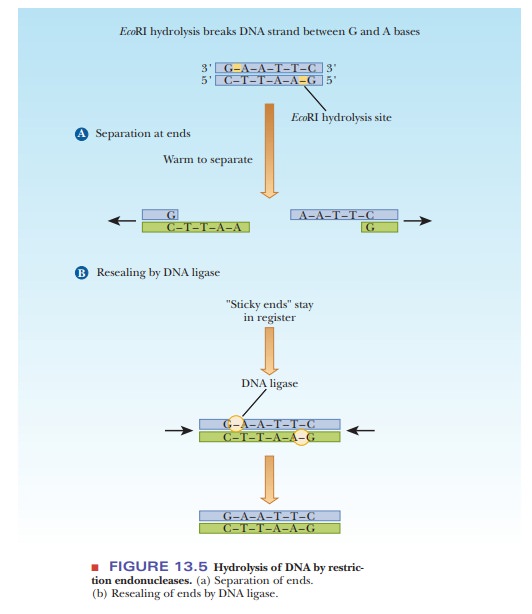
To make
life more challenging for molecular biologists, some enzymes can also cut with
less than absolute specificity. This is called star (*) activity and can often be seen if the enzyme concentration
is too high or the enzyme is incubated with the DNA too long.
Why are sticky ends important?
As we will see in the next section, the production of sticky ends by a restriction enzyme is very important to the process of creating recombinant DNA. Once DNA is cut with a particular enzyme, the sticky ends fit back together. At that point it does not matter whether the pieces came from the original piece of DNA. In other words, pieces of DNA from different sources can be put together as long as they were both cut with the same restriction enzyme.
Summary
Restriction endonucleases are enzymes produced
by bacteria that hydro-lyze the phosphodiester backbone of DNA at specific
sequences.
The sequences targeted by
restriction endonucleases are palindromes, meaning their sequence reads the
same on both strands going in the same direction.
Most
restriction enzymes cut DNA in a way that leaves sticky ends that are very
useful for recombining DNA from different sources.
Related Topics