Chapter: Biochemistry: Nucleic Acid Biotechnology
DNA Fingerprinting
DNA Fingerprinting
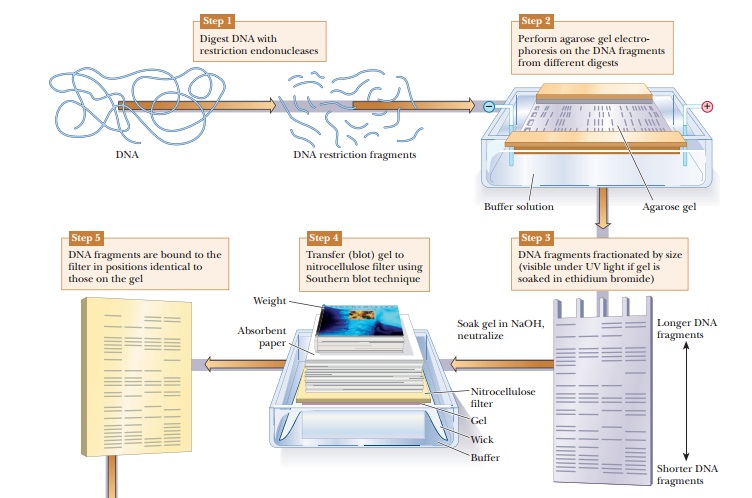
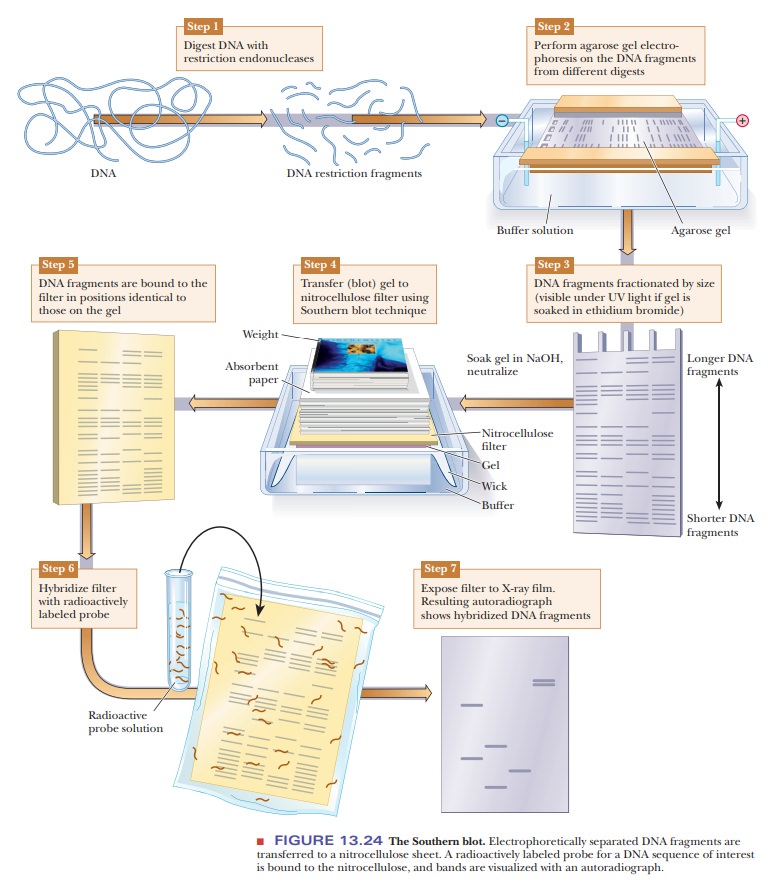
DNA samples can be studied and compared using a technique called DNAfingerprinting. The DNA is digested with restriction enzymes and then run onan agarose gel (Figure 13.24). The DNA fragments can be seen directly on the gel if it is soaked in ethidium bromide and viewed under ultraviolet light. As shown in Figure 13.24, Step 3, this gives bands of varying sizes, depending on the nature of the DNA and the restriction enzymes used. If greater sensitivity is needed, or if the number of fragments would be too great to distinguish the bands, this technique can be modified to visualize only selected DNA sequences. The first step would be to transfer the DNA to a nitrocellulose membrane in a procedure called a Southern blot, after its inventor, E. M. Southern. The agarose gel is soaked in NaOH to denature the DNA because only single-stranded DNA binds to the nitrocellulose. The membrane is placed on the agarose gel, which is on top of a filter-paper wick placed in buffer. Dry absorbent paper is placed on top of the nitrocellulose. Wicking action carries the buffer from the buffer chamber up through the gel and nitrocellulose into the dry paper. The DNA bands move out of the gel and stick to the nitrocellulose. The next step is to visualize the bands on the nitrocellulose. A specific DNA probe that is labeled with 32P is incubated with the nitrocellulose membrane. The DNA probe binds to DNA fragments that are complementary. The membrane is then placed on photographic paper to produce an autoradiograph. The use of the specific probe greatly reduces the number of bands seen and isolates desired DNA sequences.
Restriction-Fragment Length Polymorphisms:
A Powerful Method for Forensic Analysis
In organisms (such as humans) with two sets of chromosomes, a given
gene on one chromosome may differ slightly from the corresponding gene on the
paired chromosome. In the language of genetics, these genes are alleles. When they are the same on the
paired chromosomes, the organism is homozygous
for that gene; when they differ, the organism is heterozygous. A difference between alleles, even a change in one
base pair, can mean that one allele has a recognition site for a restriction
endonuclease and the other does not. Restriction fragments of different sizes
are obtained on treatment with the endonuclease (Figure 13.25); they are called
restriction-fragment length
polymorphisms, or RFLPs (pronounced
“riflips”) for short.
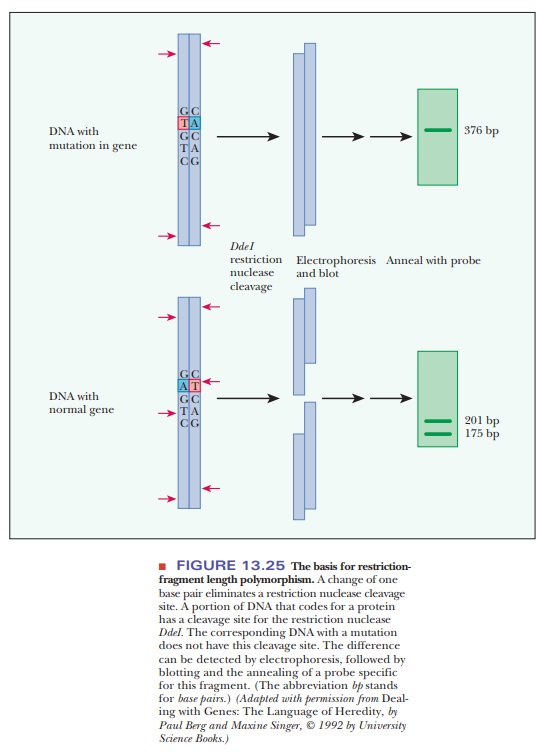
These polymorphisms (a
word meaning “many shapes”) are analyzed via gel electrophoresis to separate
the fragments by size, followed by blotting and the annealing of a probe for a
specific sequence.
How can differences in DNA from individuals be seen?
Research has shown that polymorphisms are quite common, much more so than the mutations in traits such as eye color and inherited diseases that were used for earlier genetic mapping. RFLPs can be used as markers for heredity in the same way as mutations in visible traits because they are inherited in the manner predicted by classical genetics (Figure 13.26). However, because they are much more abundant than mutations that lead to phenotypic variations, they have provided many more markers for detailed genetic mapping.
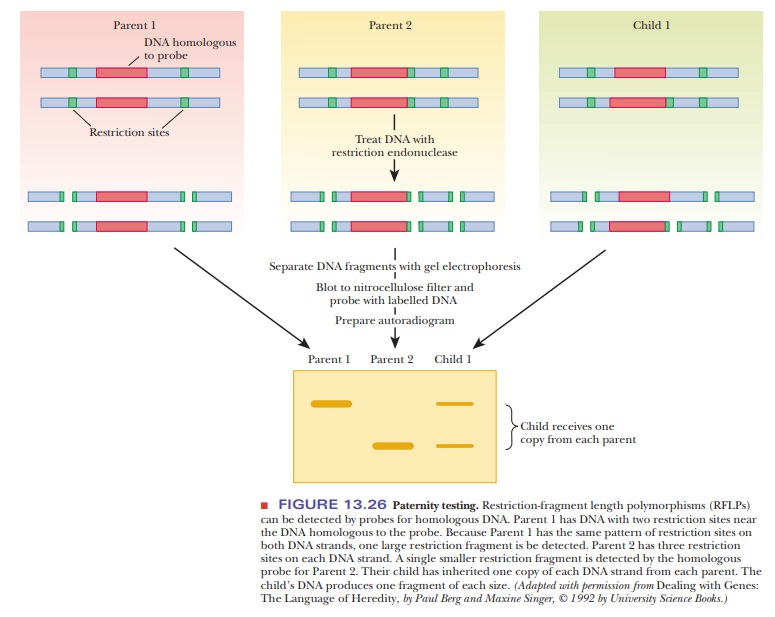
Figure 13.26 shows how RFLP analysis can be used in paternity testing. A child
gets one of each allele from each parent, so every fragment that the child has
must also be present in one of the parents. Thus, it would be easy to eliminate
a prospective father undergoing a test for paternity; if the child had a RFLP
band that neither the mother nor the prospective father has, then the
prospective father is excluded. The same kinds of analyses are done with
evidence found at crime scenes. Suspects in criminal cases may be exonerated if
their DNA samples do not match those found at the scene. RFLP analysis was used
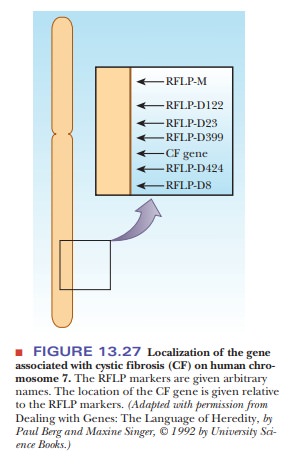
extensively in the process of locating the altered gene that causes cystic
fibrosis, a prevalent genetic disease. Once the gene was located on chromosome
7, a series of RFLP markers was used to help map its exact position (Figure
13.27). Then the gene was isolated from restriction endonuclease digests and
cloned, and its protein product was then characterized. The protein in question
is involved in the transport of chloride ion (Cl–)
through membranes. If this protein is defective, chloride ions remain in the
cells and take up water by osmosis from the surrounding mucus. The mucus
thickens as a result. In the lungs, the thickened mucus favors infections,
particularly pneumonia. The results of this disease can be tragic, leading to a
short life span in those who are affected by it. This information deepens our
insight into the nature of cystic fibrosis and provides approaches to new
treatment.
Related Topics