Chapter: Biochemistry: Nucleic Acids
Structure and types of RNA
Structure of RNA
RNAs are present in the nucleus, ribosomes and
cytoplasm of eukaryolic cells. They are involved in the transfer and expression
of genetic information. They act as primer for DNA formation. Some act as
enzymes and as coenzymes. RNA also function as genetic material for viruses.
RNAs are also polynucleotides. In RNA polymer,
purine and pyrimidine nucleotides are linked together through phosphodiester
linkages. The sugar present is ribose. The nitrogenous bases present in RNA are
adenine and guanine (purine bases), uracil and cytosine (pyrimidine bases). The
nucleotides present in RNA are adenylic acid, quanidylic acid, cytidylic acid
and uridylic acid.
Types of RNA
There are mainly three types of RNAs in all
prokaryotic and eukaryotic cells. They are (1) Messenger RNA (mRNA) 2) Transfer
RNA (tRNA) 3) Ribosomal RNA (rRNA). They differ from each other by size,
formation and stability.
1. Messenger RNA
It accounts for 1-5% of cellular RNA. They have
a primary structure. They are single standed linear molecules. They consist of
1000-10,000 nucleotides. They have a free or phosphorylated 3’ and 5’ end. They
have different life span ranging from few minutes to days.
mRNA molecules are capped at 5’ end by
methylated guanine triphosphate. Capping protects mRNA from nuclease attack. At
3’ end a polymer of adenylate (poly A) is found as the tail. Poly A tail
protects mRNA from nuclease attack.
Intrastrand base pairing among complementary
bases allows folding of the linear molecule. As a result, haripin or loop like
secondary structure is formed.
Functions
·
mRNA is
a direct carrier of genetic information from the nucleus tothe cytoplasm.
·
It
contain information required for the synthesis of protein molecules.
2. Transfer RNA
It accounts for 10-15% of total cell RNA. They
are the smallest of all the RNAs. Usually they consist of 50-100 nucleotides.
They are single standard molecules. They contain unusual bases such as
methylated adenine, guanine, cytosine and thymine,dihydrouracil and
pseudouridine. These unusual bases are important for binding 6-RNA to intra
chain base pairing. Further some bases are not involved in base pairing,
resulting in loops and arms formation in tRNA. These folding in the primary
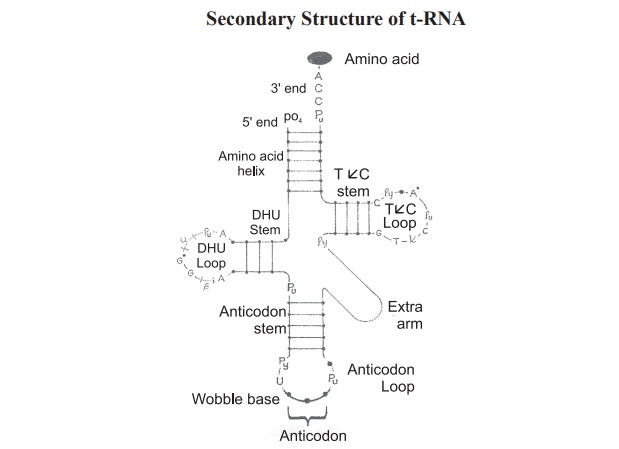
structure generates a secondary structure (Fig.7.8).
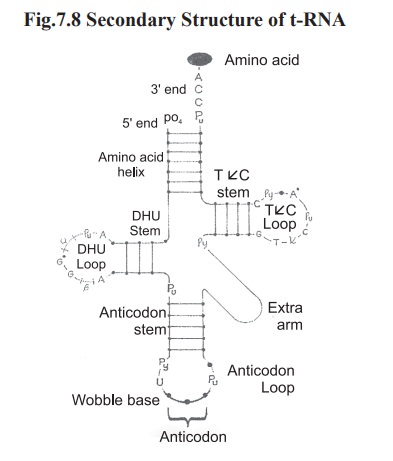
Secondary structure of t-RNA is in the form of
a clover leaf. The important feature of the clover-leaf structure are,
1.
An
acceptor arm with base sequence “CCA” 3’-OH of adenosine moiety of t-RNA.
2.
An
anticodon arm which recognises codon on mRNA.
3.
TφC arm
which contain unusual base cytosine.
4.
D- arm
which contain many dihydrouracil residues.
Functions
It is the carrier of amino acids to the site of
protein synthesis.
There is at least one t-RNA molecule to each of
20 amino acids required for protein synthesis.
3. Ribosomal RNA
This accounts for 80% of the total cellular
RNA. It is present in ribosomes. In ribosomes, r-RNA is found in combination
with protein. It is known as ribonucleoprotein. The length of rRNA ranges from
100-600 nucleotides. rRNA molecules have a secondary structure. Intra strand
base pairing between complementary bases generate double helical segments or
loops.
Functions
1.
They are
required for the formation of risosomes
2.
They are
involved in the initiation of protein synthesis.
Related Topics