Chapter: Biochemistry: Nucleic Acids: How Structure Conveys Information
How does ribosomal RNA combine with proteins to form the site of protein synthesis?
How does ribosomal RNA combine
with proteins to form the site of protein synthesis?
In
contrast with tRNA, rRNA molecules tend to be quite large, and only a few types
of rRNA exist in a cell. Because of the intimate association between rRNA and
proteins, a useful approach to understanding the structure of rRNA is to
investigate ribosomes themselves.
The RNA
portion of a ribosome accounts for 60%–65% of the total weight, and the protein
portion constitutes the remaining 35%–40% of the weight. Dissociation of
ribosomes into their components has proved to be a useful way of studying their
structure and properties. A particularly important endeavor has been to
determine both the number and the kind of RNA and protein mol-ecules that make
up ribosomes. This approach has helped elucidate the role of ribosomes in
protein synthesis. In both prokaryotes and eukaryotes, a ribosome consists of
two subunits, one larger than the other. In turn, the smaller subunit consists
of one large RNA molecule and about 20 different proteins; the larger subunit
consists of two RNA molecules in prokaryotes (three in eukaryotes) and about 35
different proteins in prokaryotes (about 50 in eukaryotes). The subunits are
easily dissociated from one another in the laboratory by lowering the Mg2+
concentration of the medium. Raising the Mg2+
concentration to its original level reverses the process, and active ribosomes
can be reconstituted by this method.
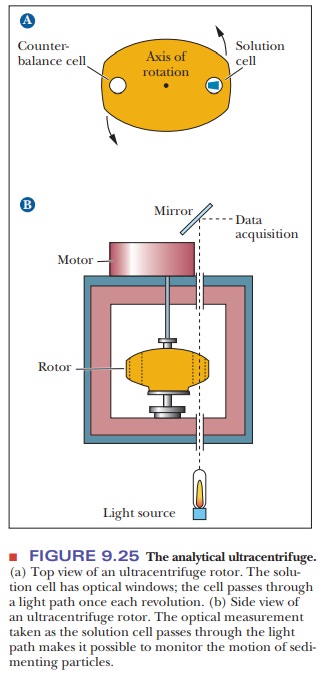
A
technique called analytical
ultracentrifugation has proved very useful for monitoring the dissociation
and reassociation of ribosomes. Figure 9.25 shows an analytical
ultracentrifuge. We need not consider all the details of this tech-nique, as
long as it is clear that its basic aim is the observation of the motion of
ribosomes, RNA, or protein in a centrifuge. The motion of the particle is
characterized by a sedimentation
coefficient, expressed in Svedberg
units (S), which are named after Theodor Svedberg, the Swedish scientist
who invented the ultracentrifuge. The S value increases with the molecular
weight of the sedi-menting particle, but it is not directly proportional to it
because the particle’s shape also affects its sedimentation rate.
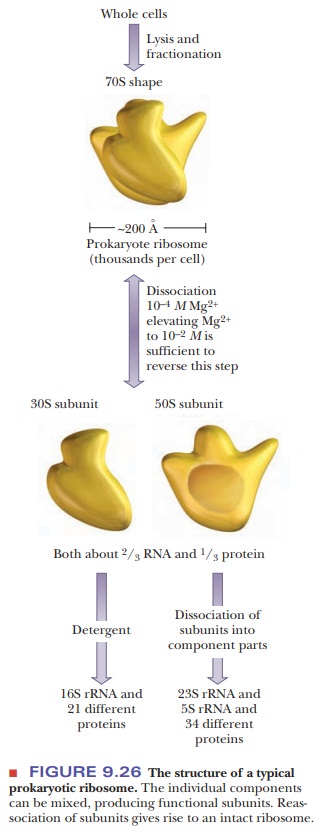
Ribosomes
and ribosomal RNA have been studied extensively via sedimen-tation
coefficients. Most research on prokaryotic systems has been done with the
bacterium Escherichia coli, which we
shall use as an example here. An E.coli ribosome
typically has a sedimentation coefficient of 70S. When an intact70S bacterial
ribosome dissociates, it produces a light 30S subunit and a heavy 50S subunit.
Note that the values of sedimentation coefficients are not addi-tive, showing
the dependence of the S value on the shape of the particle. The 30S subunit
contains a 16S rRNA and 21 different proteins. The 50S subunit contains a 5S
rRNA, a 23S rRNA, and 34 different proteins (Figure 9.26). For comparison,
eukaryotic ribosomes have a sedimentation coefficient of 80S, and the small and
large subunits are 40S and 60S, respectively. The small subunit of eukaryotes
contains an 18S rRNA, and the large subunit contains three types of rRNA
molecules: 5S, 5.8S, and 28S.
The 5S
rRNA has been isolated from many different types of bacteria, and the
nucleotide sequences have been determined. A typical 5S rRNA is about 120
nucleotide residues long and has a molecular mass of about 40,000 Da. Some
sequences have also been determined for the 16S and 23S rRNA mol-ecules. These
larger molecules are about 1500 and 2500 nucleotide residues long,
respectively. The molecular mass of 16S rRNA is about 500,000 Da, and that of
23S rRNA is about one million Da. The degrees of secondary and ter-tiary
structure in the larger RNA molecules appear to be substantial.
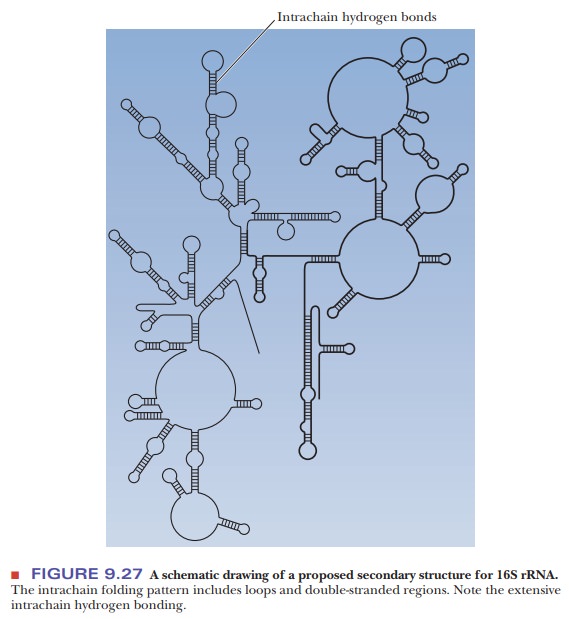
A
secondary structure has been proposed for 16S rRNA (Figure 9.27), and
suggestions have been made about the way in which the proteins associate with
the RNA to form the 30S subunit.
The self-assembly of ribosomes takes place
in the living cell, but the process can be duplicated in the laboratory.
Elucidation of ribosomal structure is an active field of research. The binding
of antibiotics to bacterial ribosomal subunits so as to prevent self-assembly
of the ribosome is one focus of the investigation. The structure of ribosomes
is also one of the points used to compare and contrast eukaryotes, eubacteria,
and archaebacteria. The study of RNA became much more exciting in 1986, when
Thomas Cech showed that certain RNA molecules exhibited catalytic activity.
Equally exciting was the recent discovery that the ribosomal RNA, and not
protein, is the part of a ribosome that catalyzes the formation of peptide
bonds in bacteria.
Related Topics