Chapter: Biochemistry: Nucleic Acids: How Structure Conveys Information
The Principal Kinds of RNA and Their Structures
The Principal Kinds of RNA and
Their Structures
What kinds of RNA play a role in life processes?
Six
kinds of RNA-transfer RNA (tRNA), ribosomal RNA (rRNA), messenger RNA (mRNA),
small nuclear RNA (snRNA), micro RNA (miRNA), and small interfering RNA (siRNA)-play
an important role in the life processes of cells. Figure 9.20 shows the process
of information transfer. The various kinds of RNA participate in the synthesis
of proteins in a series of reactions ultimately directed by the base sequence
of the cell’s DNA. The base sequences of
all types of RNA are determined by that ofDNA. The process by which the
order of bases is passed from DNA to RNA is calledtranscription.
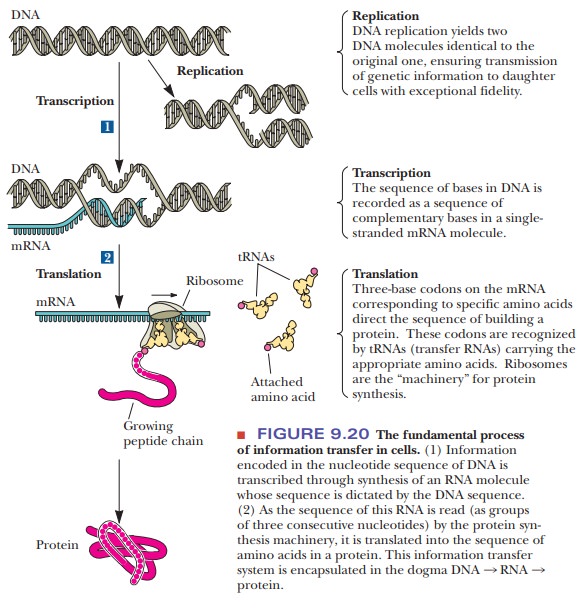
Ribosomes,
in which rRNA is associated with proteins, are the sites for assembly of the
growing polypeptide chain in protein synthesis. Amino acids are brought to the
assembly site covalently bonded to tRNA, as aminoacyltRNAs. The order of bases
in mRNA specifies the order of amino acids in the growing protein; this process
is called translation of the genetic
message. A sequence of three bases in mRNA directs the incorporation of a
particular amino acid into the growing pro-tein chain. We are going to see that
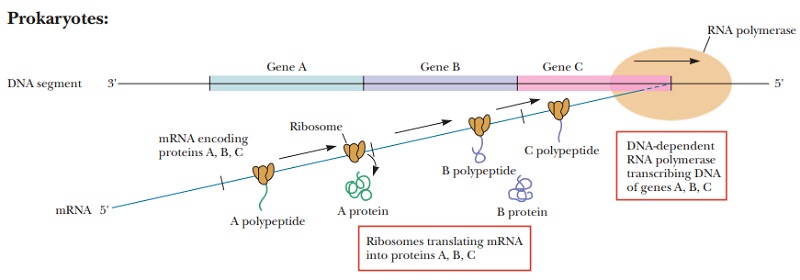
the details of the process differ in prokaryotes and in eukaryotes (Figure
9.21). In prokaryotes, there is no nuclear membrane, so mRNA can direct the
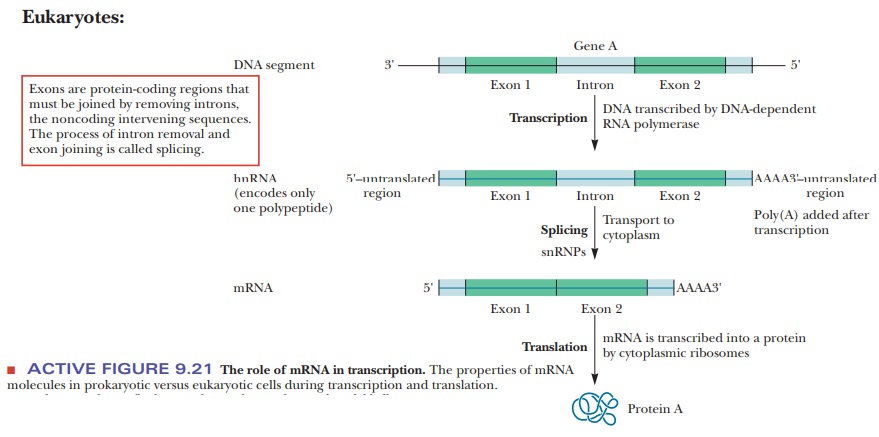
synthesis of proteins while it is still being transcribed. Eukaryotic mRNA, on
the other hand, undergoes considerable processing. One of the most important
parts of the process is splicing out intervening sequences (introns), so that
the parts of the mRNA that will be expressed (exons) are contiguous to each
other.
Small
nuclear RNAs are found only in the nucleus of eukaryotic cells, and they are
distinct from the other RNA types. They are involved in processing of initial
mRNA transcription products to a mature form suitable for export from the
nucleus to the cytoplasm for translation. Micro RNAs and small interfering RNAs
are the most recent discoveries. SiRNAs are the main players in RNAinterference (RNAi), a process that
was first discovered in plants and later inmammals, including humans. RNAi
causes the suppression of certain genes. It is also being used extensively by
scientists who wish to eliminate the effect of a gene to help discover its
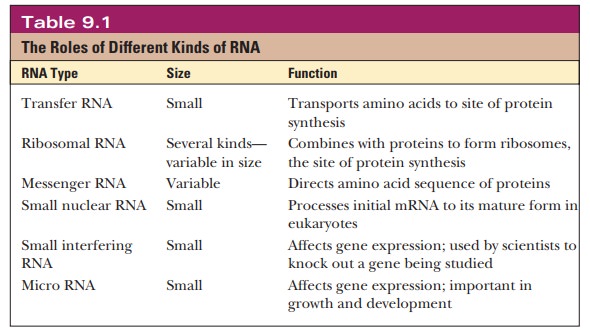
function. Table 9.1 summarizes the types of RNA.
What is the role of transfer RNA in protein synthesis?
The
smallest of the three important kinds of RNA is tRNA. Different types of tRNA
molecules can be found in every living cell because at least one tRNA bonds
specifically to each of the amino acids that commonly occur in proteins.
Frequently
there are several tRNA molecules for each amino acid. A tRNA is a
single-stranded polynucleotide chain, between 73 and 94 nucleotide residues
long, that generally has a molecular mass of about 25,000 Da. (Note that
bio-chemists tend to call the unit of atomic mass the dalton, abbreviated Da.)
Intrachain
hydrogen bonding occurs in tRNA, forming A–U and G–C base pairs similar to
those that occur in DNA except for the substitution of uracil for thymine. The
duplexes thus formed have the A-helical form, rather than the B-helical form,
which is the predominant form in DNA. The molecule can be drawn as a cloverleaf structure, which can be
considered the secondary structure of tRNA because it shows the hydrogen
bonding between
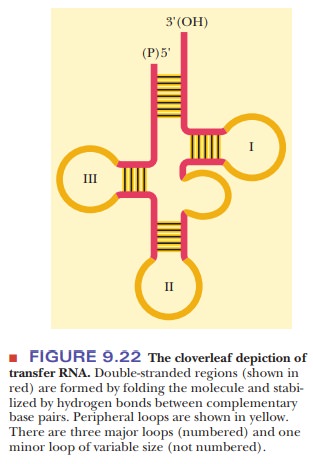
The hydrogen-bonded portions of the molecule are called stems, and the non-hydrogen-bonded
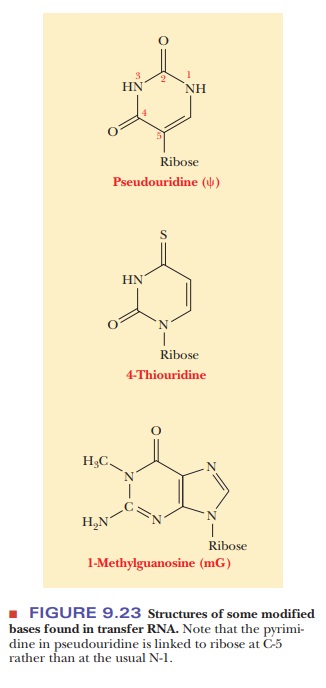
portions are loops. Some of these
loops contain modified bases (Figure 9.23). During protein synthesis, both tRNA
and mRNA are bound to the ribosome in a definite spatial arrange-ment that
ultimately ensures the correct order of the amino acids in the grow-ing
polypeptide chain.
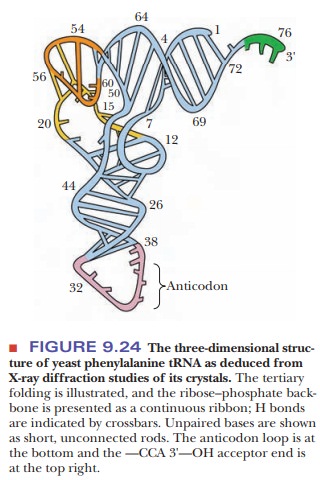
A
particular tertiary structure is necessary for tRNA to interact with the enzyme
that covalently attaches the amino acid to the 2' or 3' end. To produce this
tertiary structure, the tRNA folds into an L-shaped conformation that has been
determined by X-ray diffraction (Figure 9.24).
How does ribosomal RNA combine with proteins to form the site of protein synthesis?
In
contrast with tRNA, rRNA molecules tend to be quite large, and only a few types
of rRNA exist in a cell. Because of the intimate association between rRNA and
proteins, a useful approach to understanding the structure of rRNA is to
investigate ribosomes themselves.
The RNA
portion of a ribosome accounts for 60%–65% of the total weight, and the protein
portion constitutes the remaining 35%–40% of the weight. Dissociation of
ribosomes into their components has proved to be a useful way of studying their
structure and properties. A particularly important endeavor has been to
determine both the number and the kind of RNA and protein mol-ecules that make
up ribosomes. This approach has helped elucidate the role of ribosomes in
protein synthesis. In both prokaryotes and eukaryotes, a ribosome consists of
two subunits, one larger than the other. In turn, the smaller subunit consists
of one large RNA molecule and about 20 different proteins; the larger subunit
consists of two RNA molecules in prokaryotes (three in eukaryotes) and about 35
different proteins in prokaryotes (about 50 in eukaryotes). The subunits are
easily dissociated from one another in the laboratory by lowering the Mg2+
concentration of the medium. Raising the Mg2+
concentration to its original level reverses the process, and active ribosomes
can be reconstituted by this method.
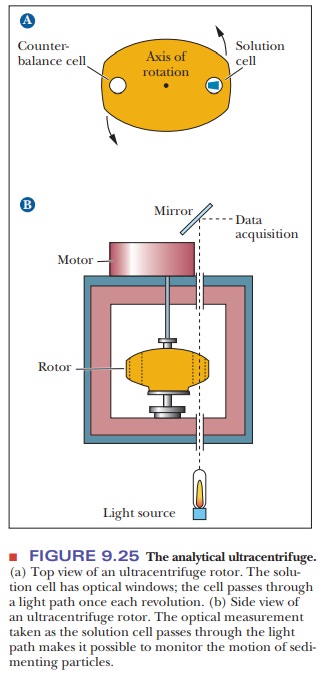
A
technique called analytical
ultracentrifugation has proved very useful for monitoring the dissociation
and reassociation of ribosomes. Figure 9.25 shows an analytical
ultracentrifuge. We need not consider all the details of this tech-nique, as
long as it is clear that its basic aim is the observation of the motion of
ribosomes, RNA, or protein in a centrifuge. The motion of the particle is
characterized by a sedimentation
coefficient, expressed in Svedberg
units (S), which are named after Theodor Svedberg, the Swedish scientist
who invented the ultracentrifuge. The S value increases with the molecular
weight of the sedi-menting particle, but it is not directly proportional to it
because the particle’s shape also affects its sedimentation rate.
Ribosomes
and ribosomal RNA have been studied extensively via sedimen-tation
coefficients. Most research on prokaryotic systems has been done with the
bacterium Escherichia coli, which we
shall use as an example here. An E.coli ribosome
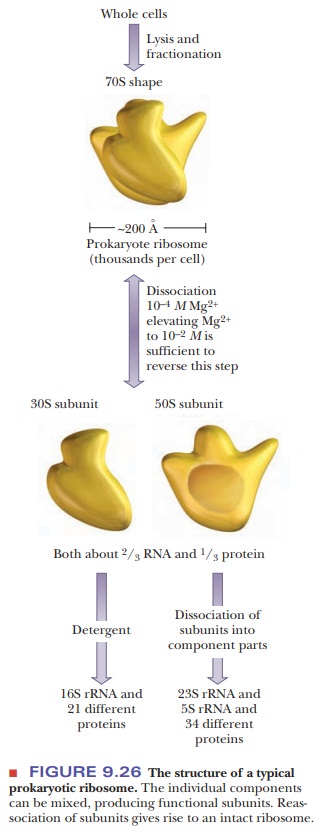
typically has a sedimentation coefficient of 70S. When an intact70S bacterial
ribosome dissociates, it produces a light 30S subunit and a heavy 50S subunit.
Note that the values of sedimentation coefficients are not addi-tive, showing
the dependence of the S value on the shape of the particle. The 30S subunit
contains a 16S rRNA and 21 different proteins. The 50S subunit contains a 5S
rRNA, a 23S rRNA, and 34 different proteins (Figure 9.26). For comparison,
eukaryotic ribosomes have a sedimentation coefficient of 80S, and the small and
large subunits are 40S and 60S, respectively. The small subunit of eukaryotes
contains an 18S rRNA, and the large subunit contains three types of rRNA
molecules: 5S, 5.8S, and 28S.
The 5S
rRNA has been isolated from many different types of bacteria, and the
nucleotide sequences have been determined. A typical 5S rRNA is about 120
nucleotide residues long and has a molecular mass of about 40,000 Da. Some
sequences have also been determined for the 16S and 23S rRNA mol-ecules. These
larger molecules are about 1500 and 2500 nucleotide residues long,
respectively. The molecular mass of 16S rRNA is about 500,000 Da, and that of
23S rRNA is about one million Da. The degrees of secondary and ter-tiary
structure in the larger RNA molecules appear to be substantial.
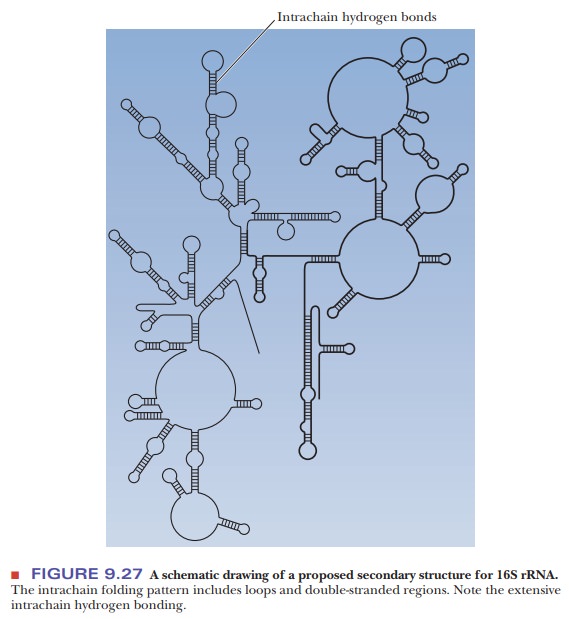
A
secondary structure has been proposed for 16S rRNA (Figure 9.27), and
suggestions have been made about the way in which the proteins associate with
the RNA to form the 30S subunit.
The self-assembly of ribosomes takes place
in the living cell, but the process can be duplicated in the laboratory.
Elucidation of ribosomal structure is an active field of research. The binding
of antibiotics to bacterial ribosomal subunits so as to prevent self-assembly
of the ribosome is one focus of the investigation. The structure of ribosomes
is also one of the points used to compare and contrast eukaryotes, eubacteria,
and archaebacteria. The study of RNA became much more exciting in 1986, when
Thomas Cech showed that certain RNA molecules exhibited catalytic activity.
Equally exciting was the recent discovery that the ribosomal RNA, and not
protein, is the part of a ribosome that catalyzes the formation of peptide
bonds in bacteria.
How does messenger RNA direct protein synthesis?
The
least abundant of the main types of RNA is mRNA. In most cells, it constitutes
no more than 5%–10% of the total cellular RNA. The sequences of bases in mRNA
specify the order of the amino acids in proteins. In rapidly growing cells,
many different proteins are needed within a short time interval. Fast turnover
in protein synthesis becomes essential. Consequently, it is logical that mRNA
is formed when it is needed, directs the synthesis of proteins, and then is
degraded so that the nucleotides can be recycled. Of the main types of RNA,
mRNA is the one that usually turns over most rapidly in the cell. Both tRNA and
rRNA (as well as ribosomes themselves) can be recycled intact for many rounds
of protein synthesis.
The
sequence of mRNA bases that directs the synthesis of a protein reflects the
sequence of DNA bases in the gene that codes for that protein, although this
mRNA sequence is often altered after it is produced from the DNA. Messenger RNA
molecules are heterogeneous in size, as are the proteins whose sequences they
specify. Less is known about possible intrachain folding in mRNA, with the
exception of folding that occurs during termination of transcription. It is
also likely that several ribosomes are associated with a single mRNA molecule
at some time during the course of protein synthesis. In eukaryotes, mRNA is
initially formed as a larger precursor molecule called heteroge-neous nuclear RNA (hnRNA). These contain lengthy portions
of interveningsequences called introns
that do not encode a protein. These introns are removed by posttranscriptional
splicing. In addition, protective units called 5'-caps and - > ' poly(A) tails are added before the mRNA
is complete.
How does small nuclear RNA help with the processing of RNA?
A
recently discovered RNA molecule is the small nuclear RNA (snRNA), which is
found, as the name implies, in the nucleus of eukaryotic cells. This type of
RNA
In the cell, it is complexed with proteins forming smallnuclear ribonucleoprotein particles, usually
abbreviated snRNPs (pronounced“snurps”).
These particles have a sedimentation coefficient of 10S. Their function is to
help with the processing of the initial mRNA transcribed from DNA into a mature
form that is ready for export out of the nucleus. In eukaryotes, transcription
occurs in the nucleus, but because most protein synthesis occurs in the
cytosol, the mRNA must first be exported.
What is RNA interference, and why is it important?
The
process called RNA interference was heralded as the breakthrough of the year in
2002 in Science magazine. Short
stretches of RNA (20–30 nucleotides long) have been found to have an enormous
control over gene expression.
This
process has been found to be a protection mechanism in many species, with the
siRNAs being used to eliminate expression of an undesirable gene, such as one
that is causing uncontrolled cell growth or a gene that came from a virus.
These small RNAs are also being used by scientists who wish to study gene
expres-sion. In what has become an explosion of new biotechnology, many
companies have been created to produce and to market designer siRNA to knock
out hun-dreds of known genes. This technology also has medical applications:
siRNA has been used to protect mouse liver from hepatitis and to help clear
infected liver cells of the disease. For the moment, we can say that many new
biotechnology companies have sprung up in recent years to exploit possible
applications of RNA interference.
Summary
Four kinds of RNA-transfer RNA, ribosomal RNA, messenger RNA, and
small nuclear RNA-are involved in protein synthesis.
Transfer RNA transports amino acids to the sites of protein
synthesis on ribosomes, which consist of ribosomal RNAs and proteins.
Messenger RNA directs the amino acid sequence of proteins. Small
nuclear RNA is used to help process eukaryotic mRNA to its final form.
RNA
interference, which requires short stretches of siRNA, controls gene
expression.
Related Topics