Chapter: Biochemistry: Nucleic Acids: How Structure Conveys Information
How does supercoiling take place in eukaryotic DNA?
How does supercoiling take place
in eukaryotic DNA?
The supercoiling of the nuclear DNA of eukaryotes (such as plants and animals) is more complicated than the supercoiling of the circular DNA from prokaryotes. Eukaryotic DNA is complexed with a number of proteins, especially with basic proteins that have abundant positively charged side chains at physiological (neutral) pH. Electrostatic attraction between the negatively charged phosphate groups on the DNA and the positively charged groups on the proteins favors the formation of complexes of this sort. The resulting material is calledchromatin. Thus, topological changes induced by supercoiling must be accommodated by the histone-protein component of chromatin.
The principal proteins in chromatin are the histones, of which there are five main types, called H1, H2A, H2B, H3, and H4. All these proteins contain large numbers of basic amino acid residues, such as lysine and arginine. In the chromatin structure, the DNA is tightly bound to all the types of histone except H1.
The H1 protein is comparatively easy to remove from chromatin, but dis-sociating the other histones from the complex is more difficult. Proteins other than histones are also complexed with the DNA of eukaryotes, but they are neither as abundant nor as well studied as histones.
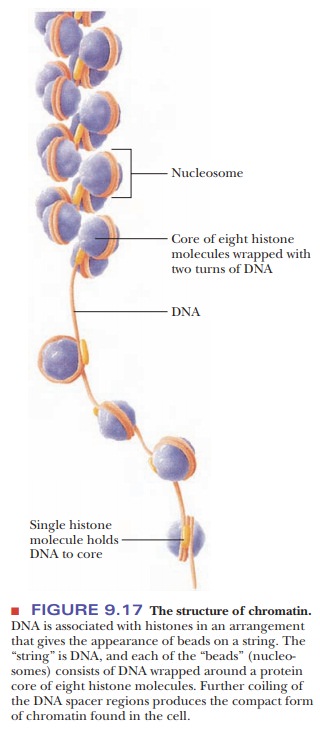
In electron micrographs, chromatin resembles beads on a string (Figure 9.17). This appearance reflects the molecular composition of the protein– DNA complex. Each “bead” is a nucleosome, consisting of DNA wrapped around a histone core. This protein core is an octamer, which includes two molecules of each type of histone but H1; the composition of the octamer is (H2A)2(H2B)2(H3)2(H4)2. The “string” portions are called spacer regions; they consist of DNA complexed to some H1 histone and nonhistone proteins. As the DNA coils around the histones in the nucleosome, about 150 base pairs are in contact with the proteins; the spacer region is about 30 to 50 base pairs long. Histones can be modified by acetylation, methylation, phosphorylation, and ubiquitinylation. Ubiquitin is a protein involved in the degradation of other proteins. Modifying histones changes their DNA and protein-binding characteristics, and how these changes affect transcription and replication is a subject of active research.
Related Topics