Chapter: Genetics and Molecular Biology: Genetic Engineering and Recombinant DNA
Isolation of DNA
The Isolation of DNA
Cellular DNA, chromosomal or nonchromosomal, is the
starting point of many genetic engineering experiments. Such DNA can be
extracted and purified by the traditional techniques of heating the cell
extracts in the presence of detergents and removing proteins by phenol
extraction. If polysaccharides or RNA contaminate the sample, they can be re-moved
by equilibrium density gradient centrifugation in cesium chloride.
Two types of vectors are commonly used: plasmids
and phage. A plasmid is a DNA element similar to an episome that replicates
inde-pendently of the chromosome. Usually plasmids are small, 3,000 to 25,000
base pairs, and circular. Generally, lambda phage or closely related
derivatives are used for phage vectors in Escherichia
coli, but for cloning in other bacteria, like Bacillus subtilis, other phage are used. In some cases a plasmid can
be developed that will replicate autonomously in more than one host organism.
These “shuttle” vectors are of special value in studying genes of eukaryotes;
we will consider them later.
Most often, complete purification of plasmid DNA is
unnecessary and useable DNA can be obtained merely by lysing the cells, partial
removal of chromosomal DNA, and the removal of most protein.
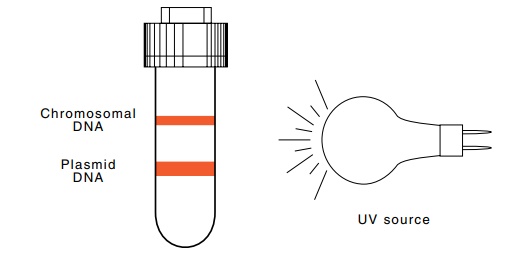
Figure 9.1
The separation of covalently
closed plasmid DNA from nickedplasmid DNA and fragments of chromosomal DNA on
CsCl equilibrium density gradient centrifugation in the presence of ethidium
bromide. The bands of DNA fluoresce upon illumination by UV light.
Intricate DNA constructions often require highly pure DNA to avoid extraneous nucleases or
inhibition of sensitive enzymes. The complete purification of plasmid DNA
generally requires several steps. After the cells are opened with lysozyme
which digests the cell wall and detergents are added to solubilize membranes
and to inactivate some proteins, most of the chromosomal DNA is removed by
centrifugation. For many purposes chromatographic methods can be used to
complete the purification. When the highest purity is required, however, the
plasmid is purified by equilibrium density gradient centrifugation. This is
done in the presence of ethidium bromide. Any chromosomal DNA remaining with
the plas-mid will have been fragmented and will be linear, whereas most of the
plasmid DNA will be covalently closed circles. As we saw, intercalating ethidium
bromide untwists the DNA. For a circular mole-cule this untwisting generates
supercoiling whereas for a linear mole - cule the untwisting has no major
effects. Therefore a linear DNA molecule can intercalate more ethidium bromide
than a circular mole-cule. Since ethidium bromide is less dense than DNA, the
linear DNA molecules with intercalated ethidium bromide “float” relative to the
circles, and therefore the two species can easily be separated. Following the
centrifugation, the two bands of DNA are observed by shining UV light on the
tube (Fig. 9.1). The natural fluorescence of ethidium bromide is enhanced 50
times by intercalation into DNA, and the bands glow a bright cherry red under
UV light.
Lambda phage may also be partially purified by rapid techniques that remove cell debris and most contaminants. A more complete purification can be obtained by utilizing their unique density of 1.5 g/cm3, which is halfway between the density of protein, 1.3, and the density of DNA, 1.7. The phage may be isolated by equilibrium density gradient centrifugation in which the density halfway between the top and bottom of the centrifuge tube is 1.5. They too may be easily observed in the centrifuge tube. They form a bluish band, which results from the preferential scattering of shorter wavelengths of light known as Tyndall effect. This same phenomenon is the reason the sky is blue and sunsets are red.
Related Topics