Chapter: Genetics and Molecular Biology: Genetic Engineering and Recombinant DNA
Joining DNA Fragments
Joining DNA Fragments
Having discussed how DNA molecules can be cut and
purified, it is now necessary to discuss the joining of DNA molecules. In vivo, the enzyme DNA ligase repairs
nicks in the DNA backbone. This activity may also be utilized in vitro for the joining of two DNA
molecules. Two require-
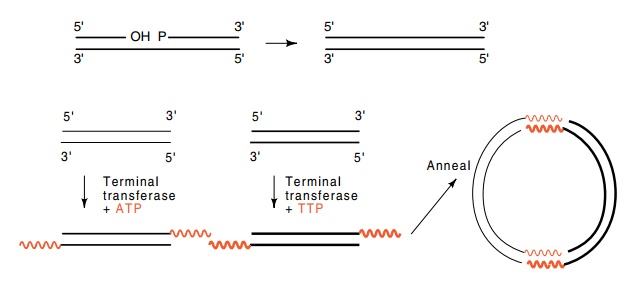
Figure
9.6 Joining two DNA fragments by
poly-dA and poly-dT tails.
ments must be met. First, the molecules must be the
correct substrates, that is, they must possess 3’-hydroxyl and 5’-phosphate groups.
Second, the groups on the molecules to be joined must be properly positioned
with respect to one another. The method for generating the proper positioning
has two variations: either to hybridize the fragments to-gether via their
sticky ends or, if flush-ended fragments are to be joined, to use such high
concentrations of fragments that from time to time they are spontaneously in
the correct positions.
Hybridizing DNA fragments that possess
self-complementary, or sticky ends, generates the required alignment of the DNA
molecules. Many restriction enzymes such as EcoRI
produce four-base sticky ends that can be ligated together after the sticky
ends of the pieces to be joined have hybridized together. Because the sticky
ends are usually just four base pairs, lowering the temperature during ligation
to about 12°C facilitates the hybridization-ligation process.
The flush ends of DNA molecules that are generated
by some restric-tion enzymes generate problems. One solution is to convert
flush-ended molecules to sticky-ended molecules by enzyme terminal transferase.
This enzyme adds nucleotides to the 3’ end of DNA. Poly-dA tails can be put on
one fragment and poly-dT tails can be added to the other fragment (Fig. 9.6).
The two fragments can then be hybridized together by virtue of their
self-complementary ends and ligated together. If the tails are long enough, the
complex can be directly introduced into cells, where the gaps and nicks will be
filled and sealed by the cellular enzymes. More commonly, the polymerase chain
reaction as described in the next would be used to generate any desired ends on
the molecules.
Flush ended molecules can also be joined directly
with DNA ligase. While this method is straightforward, it suffers from two
drawbacks: It requires high concentrations of DNA and ligase for the reaction
to proceed, and even then the ligation efficiency is low. Also, it is difficult
later to excise the fragment from the vector.
Linkers can also be used to generate
self-complementary single-stranded molecules (Fig. 9.7). Linkers are short,
flush-ended DNA molecules containing the recognition sequence of a restriction
enzyme that produces sticky ends. The ligation of linkers to DNA fragments
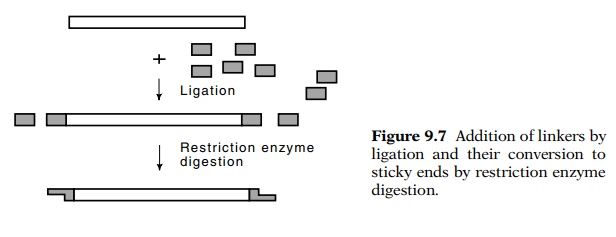
proceeds with reasonably high efficiency because
high molar concen-trations of the linkers may easily be obtained. After the
linkers have been joined to the DNA segment, the mixture is digested with the
restriction enzyme, which cuts the linkers and generates the sticky ends. In
this way a flush-ended DNA molecule is converted to a sticky-ended mole-cule
that may easily be joined to other DNA molecules.
Related Topics