Chapter: Genetics and Molecular Biology: Genetic Engineering and Recombinant DNA
Enzymatic DNA Sequencing
Enzymatic DNA Sequencing
Sanger and his colleagues developed an enzymatic
method for generating the set of DNA fragments necessary for sequence
determination. Utilization of biological tricks now makes this method
particularly efficient. When coupled with techniques to be discussed in the
next, thousands of nucleotides per day per person can be sequenced. The Sanger
method relies on the fact that elongation of a DNA strand by DNA polymerase
cannot proceed beyond an incorporated 2’,3’-dide-oxyribonucleotide. The absence
of the 3’-OH on such an incorporated nucleotide prevents chain extension.
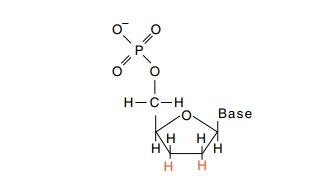
A growing chain therefore terminates with the incorporation of a
dideoxyribonucleotide. Thus, elonga-tion reactions are performed in the
presence of the four deoxyribonu-cleotide triphosphates plus sufficient
dideoxyribonucleotide triphosphate that about one of these nucleotides will be
incorporated per hundred normal nucleotides. The DNA synthesis must be
performed so that each strand initiates from the same nucleotide and from the
same strand. This is accomplished first by using only a single-stranded
tem-plate and second by hybridizing an oligonucleotide to the DNA to provide
the priming 3’-OH necessary for DNA elongation by DNA polymerases.
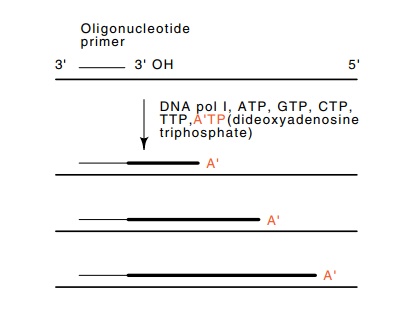
Figure
9.20 The Sanger sequencing method.
Primer extension by DNA pol Iin the presence of dideoxyadenosine triphosphate,
A’TP, creates a nested set of oligomers ending in A’.
In sequencing, four different elongation reactions
are performed, each containing one of the four dideoxyribonucleotides.
Radioactive label is introduced to the fragments by the use of either labeled
nucleo-tides or labeled primer. The fragments generated in the elongation
reactions are analyzed after electrophoresis in the same manner as the
fragments generated by chemical cleavage (Fig. 9.20).
Viruses like M13 and f1 that encapsidate only one
strand are one source of pure single-stranded DNA. Purification of this
material from as little as 2 milliliters of growth medium yields sufficient DNA
for sequencing. M13 has no rigid structural limitations on the length of viral
DNA that can be encapsidated because its DNA is packaged as a rod during its
extrusion through the cell membrane. Therefore large and small fragments may be
inserted into nonessential regions of the virus genome. Although the viral form
of the virus is single-stranded, the genetic engineering of cutting and
ligating is done with the double-stranded intracellular replicative form of the
virus DNA. This DNA can then be transformed into cells just as plasmid DNA is.
Instead of cloning the DNA to be sequenced into the
virus, it is simpler to clone the virus replication origin into a plasmid
vector that contains a nonviral origin as well. Then, cloning and normal
genetic engineering operations can be performed on the plasmid DNA, and when
sequencing is to be done, the cells can be infected with virus. Under the
influence of the virus encoded proteins, the viral replication origin on the
plasmid directs the synthesis of plus strand DNA. This is then isolated and
used in sequencing or purified and used in other operations.
The single-stranded DNA template necessary for
Sanger sequencing may also be obtained by denaturing double-stranded DNA and
prevent-ing its rehybridization. Because partial rehybridization, often not in
register, can still occur in many positions, this method works best with the
use of a DNA polymerase capable of replicating through such
obstacles. A DNA polymerase encoded by phage T7 is
helpful in this regard. Either a chemical modification of the wild-type enzyme
or a special mutant enzyme makes the T7 DNA polymerase highly processive and
capable of replicating through regions containing significant amounts of
secondary structure. The DNA pol I from E.
coli or the Klenow fragment of this enzyme are adequate for sequencing from
pure single-stranded DNA.
Needless to say, the vast explosion in the amount
of sequenced DNA has stimulated development of highly efficient methods for
analysis. Computer programs have been written for comparing homology be-tween
DNA sequences, searching for symmetrical or repeated se-quences, cataloging
restriction sites, writing down the amino acid sequences resulting when the DNA
is transcribed and translated into protein, and performing other time-consuming
operations on the se-quence. The amount of information known about DNA and the
ease in manipulating it has greatly extended our understanding of many
bio-logical processes and is now becoming highly important in industrial and
medical areas as well.
Related Topics