Chapter: Genetics and Molecular Biology: Genetic Engineering and Recombinant DNA
Biology of Restriction Enzymes
The Biology of Restriction Enzymes
In this section we digress into the
biology of restriction enzymes and then return to their use in cutting DNA. A
large number of enzymes have now been found that cut DNA at specific sites. For
the most part the enzymes come from bacteria. These enzymes are called
restriction enzymes because in the few cases that have been carefully studied,
the DNA cleaving enzyme is part of the cell’s restriction-modification sys-tem.
The
phenomenon of restriction-modification in bacteria is a small-scale immune
system for protection from infection by foreign DNA. In contrast to higher
organisms in which identification and inactivation of invading parasites,
bacteria, or viruses can be performed extracellularly, bacteria can protect
themselves only after foreign DNA has entered their cytoplasm. For this
protection, many bacteria specifically mark their own DNA by methylating bases
on particular sequences with modifying enzymes. DNA that is recognized as
foreign by its lack of methyl groups on these same sequences is cleaved by the
restriction enzymes and then degraded by exonucleases to nucleotides. Less than
one phage out of 104 wrongly methylated infecting phage is able to
grow and lyse an E. coli protected by some
restriction-modification systems. Bacteria fur-ther protect themselves from
plant and animal DNA. Much plant and
animal DNA is methylated on the cytosine in CpG sequences. Many strains of
bacteria also contain enzymes that cleave DNA when it is methylated on specific
positions.
Arber studied restriction of lambda phage in E. coli
and found that E. coli strain C does not contain a restriction-modification
system.Strain B has one restriction-modification system, and yet a different
one recognizes and methylates a different nucleotide sequence in strain K-12.
Phage P1 also specifies a restriction-modification system of its own, and this
can be superimposed on the restriction-modification system of a host in which
it is a lysogen.
Table 9.1
Plating Efficiencies of Phage
Grown onE.coli C, K, and B and Plated
on These Bacteria
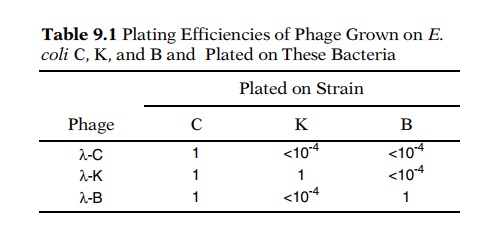
Let the notation λ-C represent lambda phage that has been grown on E. coli strain C.
Infection of strains B, K-12, and C withλgrown on thevarious strains yields different efficiencies of plaque
formation (Table 9.1). Passage of the phage through a host of one type modifies
the DNA so that it is recognized as “self” and plates at high efficiency if the
phage reinfects that same strain. It is recognized as “foreign” and plates at
low efficiency if the phage infects a strain with a different
restriction-modification system.
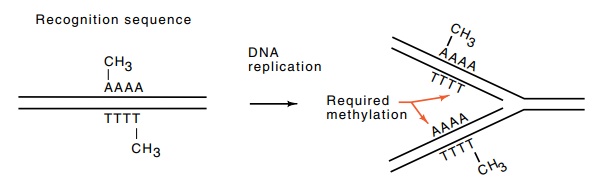
Figure 9.2 Methylation of an asymmetrical sequence necessitates recognitionand methylation of two different sequences on the daughter strands on DNA replication.
Possession of a restriction-modification system
introduces complexities to the process of DNA replication. Imagine that the
double-stranded DNA contains methyl groups on both strands of the DNA at a
recognition sequence. DNA replication creates a new duplex in which one of the
strands in each of the daughter duplexes at first lacks the modification. This
half-methylated DNA must not be recognized as foreign DNA and cleaved, but
instead must be recognized as “self” and methylated (Fig. 9.2). Therefore, the
restriction-modification system functions like a microcomputer, recognizing
three different states of methylation of its recognition sequence and taking
one of three different actions. If the sequence is unmethylated, the enzymes
cleave. If the DNA is methylated on one of the two strands, the modification
system methylates the other strand; if the DNA is methylated on both strands,
the enzymes do nothing.
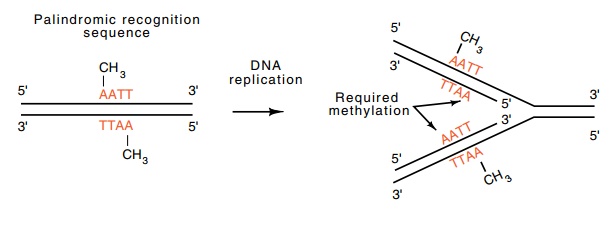
A palindromic recognition sequence streamlines
operation of the restriction-modification system. A palindrome is a sequence
that reads the same forward and backward, such as repaper and radar.
Because DNA strands possess a direction, we consider a DNA sequence to be
palindromic if it is identical when read 5’ to 3’ on the top strand and on the
bottom strand (Fig. 9.3). Palindromes, of course, can be of any size, but most
that are utilized as restriction-modification recognition se-quences are four,
five, six, and rarely, eight bases long. By virtue of the properties of
palindromes, both daughter duplexes of replicated palin-dromic sequences are
identical, and thus the modification enzyme needs to recognize and methylate
only one type of substrate (Fig. 9.4). As we already saw in Fig. 9.2, the use
of nonpalindromic recognition sequences would require that the modification
enzyme recognize two

Restriction
enzymes are divided into three main classes. The enzymes in class I form a
complex consisting of a cleaving subunit, a methylating subunit, and a sequence
recognition subunit. These enzymes cleave at sites far distant from their
recognition sequences and will not be further discussed here even though they
were the first to be discovered. Those in class II possess their sequence
recognition and cleaving activities together. They cleave in or near their
recognition sequence and are of the most use in genetic engineering. The class
III enzymes possess a cleavage subunit associated with a recognition and
methylation subunit. These cleave near their recognition site.
A
restriction enzyme within a cell is a time bomb because physical-chemical
principles limit the enzyme’s specificity for binding. If a restriction enzyme
were to bind to a wrong sequence, and a typical bacterium contains about 4 × 106 such sequences,
most likely the sequence would not be methylated and the enzyme could cleave.
This would break the chromosome, and the cell would die. The experimental
observation, however, is that cells containing restriction enzymes do not
noticeably die any faster than cells without restriction enzymes. How, then, is
the extraordinarily high specificity of the restriction enzymes generated?
Figure
9.4 Methylation of a palindromic
sequence permits recognition andmethylation of only one sequence during DNA
replication.
The requisite high specificity can be obtained if
cutting the DNA duplex is a two-step process. An enzyme could bind to the
recognition sequence, cleave one strand, wait a while, then cleave the other
strand. This has the effect of utilizing the recognition sequence twice for
each cleavage. If the enzyme binds at a site other than the recognition
sequence, it rapidly dissociates before cleaving the second strand. Therefore,
restriction enzymes are likely to produce nicks in the DNA at sites other than
the recognition sequence, but these nicks can be repaired with DNA ligase and
the cell will not be harmed in the process. Few restriction enzymes are likely
to be found that cleave both strands of the DNA at an incorrect site in a
concerted process.
Related Topics