Chapter: Biotechnology Applying the Genetic Revolution: Pathway Engineering
Biorefining of Fossil Fuels
BIOREFINING
OF FOSSIL FUELS
The growth of industrial civilization, in particular the use of fossil fuels for energy and the development of the organic chemical industry, have led to the pollution of the environment with a wide range of compounds of nonbiological origin.
Many fossil fuel deposits, of
both coal and oil, contain a high percentage of sulfur—up to 5% sulfur for many
coals from Eastern Europe or the American Midwest. Burning high- sulfur coal
releases large quantities of sulfur dioxide into the atmosphere, which leads to
the formation of acid rain. Among the possible solutions to this problem is to
develop bacteria capable of removing the offending sulfur compounds from the
coal (or oil) before combustion.
Several naturally occurring
sulfur bacteria such as Thiobacillus
and Sulfolobus can convert pyrites
(FeS2) the major form of inorganic sulfur found in coal, into
soluble sulfate that can be rinsed away. The crucial issue, therefore, is to
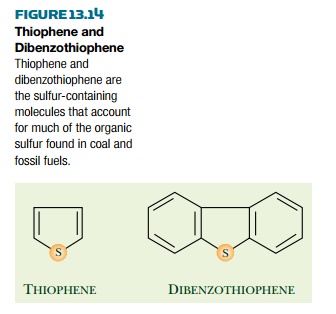
remove the organic sulfur, especially that found in thiophene rings, which typically accounts for 70% or more of the
organic sulfur (Fig. 13.14). Although compounds containing thiophene rings are
almost never found in modern-day living organisms, they form a substantial part
of the organic sulfur fraction of fossil fuels such as coal and oil. The major
quinone of Archaebacteria such as Sulfolobus
is caldariellaquinone, which contains a thiophene ring fused to a
benzoquinone. Conceivably, the fused
thiophenes of coal are the metabolic fossils of archaebacterial metabolism.
Dibenzothiophene (DBT) is a widely used model compound (Fig. 13.14) thought
to be representative of the organic
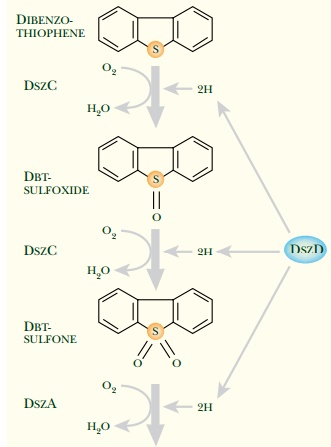
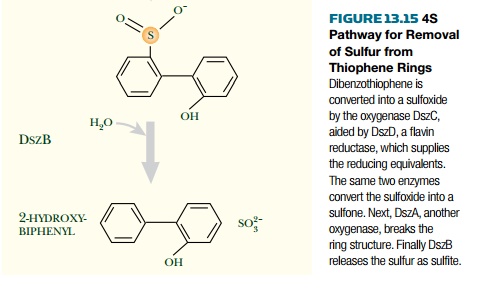
sulfur in coal and oil. Biodegradation of DBT and removal of sulfur involves
several steps, a scheme known as the 4S
pathway (Fig. 13.15). Most bacteria capable of degrading thiophene
derivatives show only partial breakdown. Many do not completely remove the
sulfur from its organically bound form, and others only use DBT that has
already been oxidized to the sulfone or sulfoxide level (see Fig. 13.15). Full
desulfurization requires
either finding a natural isolate that can carry out all the steps or the
genetic assembly of the individual steps of the 4S pathway from different
bacteria into a final engineered strain.
Certain bacteria, especially
certain species of Rhodococcus, do indeed completely desulfurize
dibenzothiophene, as well as degrade related heterocyclics such as dibenzofuran
and xanthones. The dszABC operon of Rhodococcus is found on a linear plasmid
and encodes three enzymes responsible for the 4S pathway. In addition a flavin
reductase encoded by the dszD gene is
needed to supply reduced FMN (see Fig. 13.15). The dszD gene is not linked to the dszABC
operon. The enzymes are as follows:
■
Third step: DszA = dibenzothiophene sulfone oxygenase
■
Fourth step: DszB = benzene sulfinate desulfinase
■
First and second steps: DszC = dibenzothiophene oxygenase
■
FMNH2 supply:
DszD = flavin reductase
However, there are several problems with these
natural isolates:
(a) Rhodococcus
is not well characterized genetically, so further modification is difficult.
(b) Desulfurizing
coal or oil will require robust bacteria that can be grown easily to high
density. Although not feeble, Rhodococcus is not especially convenient in this
respect.
(c) The
desulfurizing genes (dszABC) in Rhodococcus are used naturally to obtain sulfur
(not to degrade benzothiophenes for carbon and energy).
Consequently, they are expressed only at a low
level for bacterial growth. Moreover, other sulfur compounds, both inorganic
and organic, repress the operon. Thus, the inorganic sulfur present in most
high-sulfur coal and oil would repress the dszABC genes. Furthermore, pregrowth
of the dibenzothiophene and related compounds. In these cases, the same enzyme
hydroxylates the aromatic rings and also adds oxygen to the thiophene sulfur,
giving the sulfoxide and then the sulfone. The DszC enzyme itself has also been
mutated to High-level operation of the
4S pathway requires a large flow of reducing equivalents. In cells carrying a
cloned dszABC operon, reduction of FMN by flavin reductase becomes the limiting
factor in removal of sulfur from dibenzothiophene. However, flavin reductases
from several other bacteria work as well as or better than the Rhodococcus DszD
enzyme. For example, the HpaC enzyme from E. coli has been cloned and expressed
at high levels, and it greatly speeds desulfurization. As a further
modification, the hpaC gene and the dszABC genes have been joined together to
form a single operon under control of the tac promoter. (This is a hybrid promoter with the RBS from the trp promoter and the operator of the lac promoter. It is therefore a strong
promoter that is induced by IPTG.) Thus the combined desulfurization module can
be induced by IPTG when required.
Despite adding oxygen to the
sulfur of dibenzothiophene, the DszC enzyme is closely related to the ring
dioxygenases that add two hydroxyl groups to aromatic rings.
Thus phenanthrene dioxygenase hydroxylates phenanthrene (a three-ringed aromatic hydrocarbon) as well as converting DBT to its sulfone. Again, mutation of biphenyl dioxygenase by a gene shuffling approach gives mutant enzymes capable of handling broaden its substrate range. For example, a Val261Phe mutation allows oxidation of methylbenzothiophene and alkyl thiophenes.
Related Topics