Chapter: Genetics and Molecular Biology: Lambda Phage Integration and Excision
Structure of the Intasome
Structure of the Intasome
One approach to the study of the integration and
excision reactions is to determine where purified Int, Xis, FIS, and IHF
proteins bind in the att regions. The
proteins have a complex binding pattern in
attP (Fig.18.18). Int protein binds to seven sites in attP: two in the common core, two on the P arm, and three on the P’
arm. Interestingly, the core sequences for Int protein binding are different
from the arm sequences. Not surprisingly then, Int protein possesses two
domains, an N-terminal domain which binds the arm sequences and the C-terminal
domain, which binds the core sequence at the crossover point. IHF binds to
three sites, Xis protein binds to two sites, and FIS binds to one site
partially overlapping an Xis site. Together these binding sites cover the
entire region from -150 to +100 with respect to the center of the common core.
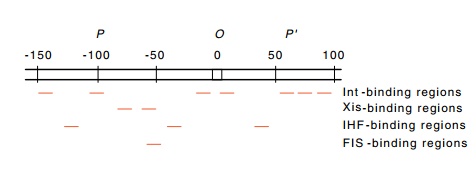
Figure 18.18 The regions inattPto which Int, Xis, FIS, and IHF proteins bind.
Several facts suggest that the protein-attP complex is folded rather than being
extended in a line. When the complex of the various att regions and the Int, Xis, FIS, and IHF proteins is examined in
the electron microscope, a compact and topologically complicated struc-ture is
seen. This would make us fear that it could be an artifact except that a
complex of Int plus Xis proteins bound at attR
appears to pair specifically with a similar complex formed at attL on a different DNA molecule. Since
the bacteria and phage att regions
possess no sequence homology except for their common core sequences, either it
is the bound proteins or it is recombined DNA that holds the pair of DNA
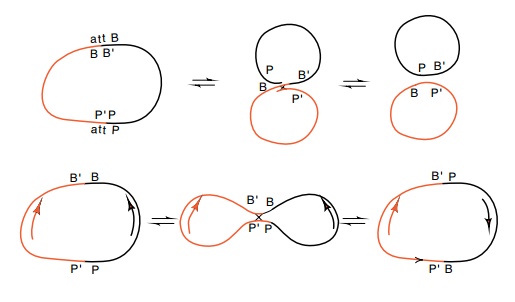
Figure
18.19 An integrative reaction between
thePP’andBB’sites in oppositeorientations on two different plasmids either
generates two smaller circles, or it inverts one of the segments between the
sites with respect to the other site.
molecules together. The attB site with which attP
interacts is much more compact, possessing only the two core binding sites of
Int protein. The attB site probably
does not bind Int protein by itself under normalphysiological conditions, but
the bare DNA merely collides with the intasome complex at attP and the strand exchange process then begins.
One way to estimate the shape of attP might be to consider the bending in
the DNA that is generated by each of the proteins which bind in the region.
Another approach for learning about the structure of the intasome is to examine
the topology of the DNA that engages in recom-bination. Consider the crossover
reaction when performed on a small circular plasmid lacking supercoiled turns.
When the att sites are in one
orientation, the crossover reaction generates two smaller circles from the
original, but when one of the att
sites is reversed, the product of a crossover reaction does not generate two
circles (Fig. 18.19). Instead, the segment of DNA between the sites is reversed
in orientation. Further, depending on the topology of the DNA at the time of
the crossover, either a simple circle or a knotted product called a trefoil is
generated (Fig. 18.20). When one of the att
sites participating in the crossover reaction is wrapped around a core of
proteins as indicated, a superhelical turn is generated. This can be trapped in
the reaction so that the product is a trefoil rather than a simple circle.
Nash and colleagues found two important results
when examining the products of Int protein-catalyzed segment reversal. First,
that half the products were trefoils, and second, that all the trefoils were
topologi-cally identical, that is, they possessed crossover points of the same
sense. The presence of only one kind of crossover points or nodes in the
trefoils means that the polarities of the wrapping in all the original PP’
sites
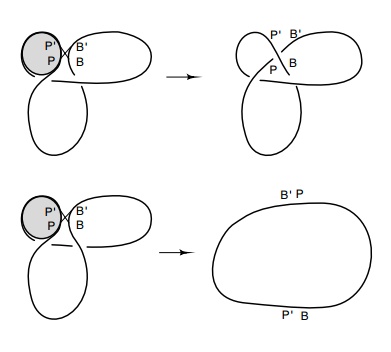
Figure
18.20 An integrativereaction between POP’ sites wrapped around a protein and BOB’ sites on a circle can generate a
trefoil if the DNA strand is trapped by the reac-tion and if the correct
strand, in this case the P’B product,
is placed above the PB’ prod-uct.
were the same. Had any been wrapped the other way
any resultant trefoils would have possessed nodes of the opposite topological
sign and would be the mirror image of the one shown. Second, the fact that the
fraction of trefoils was as large as one half means that almost all of the attP substrate was wrapped around
protein at the time of recombination.
Related Topics