Chapter: Genetics and Molecular Biology: Lambda Phage Integration and Excision
In vitro Assay of Integration and Excision
In vitro Assay of Integration and Excision
Although
ingenious genetic experiments can reveal much about biological systems,
ultimately the details must be studied biochemically. There-fore it was of
great interest to isolate the Int and Xis proteins. Such an isolation requires
an assay for the proteins. The most reliable assay for Int protein activity is
to seek a protein that carries out the entire integration reaction in vitro. As mentioned above, one likely
require-ment for such a reaction to proceed in
vitro is a high concentration of attB
and attP regions. The easiest way
to obtain these is to place bothon the same DNA molecule.
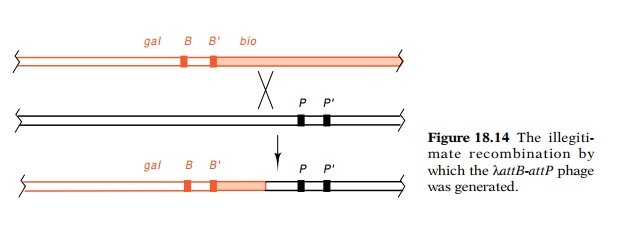
Can attB and attP be put on the same molecule, just as attL and attR could be
put on λatt2? Nash
constructed such phage by selecting the product of a rare recombination event
in a region of nonhomology between a λdgal-bio
containing attB and a λ (Fig. 18.14). Similar to the
excision-type of reaction that occurs on λatt2, on λattB-attP an
integra-tion-type of reaction can occur. This removes the bio region and leaves the phage considerably more resistant to Mg++
chelation or heat. Hence, the parental phage and derivatives that have
undergone an integration reaction and have become smaller can easily be
distinguished.
Initial tests with the λattB-attP phage showed that an in vitro integra-tion reaction catalyzed by an extract from cells would work. The experiment
was performed by incubating the λattB-attP DNA with
a cell extract prepared from heat-pulsed lambda lysogens. Then the DNA was
extracted from the mix and used to transfect cells that had been made capable
of taking up naked lambda DNA. The phage from the trans-fected spheroplasts
were spread on a lawn of sensitive cells on a plate containing pyrophosphate.
Under these conditions, only products from the integrative reaction would
produce plaques. This provided an ex-tremely sensitive assay for the
integration reaction. Later, as the assay conditions were improved, the
integration reaction could be assayed merely by physical quantitation of the
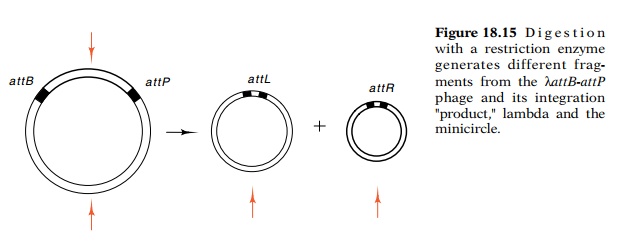
DNA products. The locations of restriction enzyme cleavage sites in the DNA are
rearranged by the integration reaction. This creates new sizes of restriction
fragments that can be detected by separating the resulting fragments by
electrophoresis (Fig. 18.15).
The in vitro integration reaction required
Mg++, ATP, and spermidine. More careful examination showed that the
use of supercoiled DNA eliminated the requirement for ATP and Mg++.
The assays permitted the purification and characterization of biologically
active Int protein, and analogous experiments have been done with the λatt2 for the
purification of Xis protein.
Related Topics