Chapter: Genetics and Molecular Biology: Transposable Genetic Elements
Structure and Properties of Tn Elements
Structure and Properties of Tn Elements
Bacterial transposons can be grouped into four broad structural classes. Members of the class containing Tn5 and Tn10 possess a pair of IS elements on either side of a DNA sequence that is transposed. Although the right- and left-hand IS elements are nearly identical, only one of the IS elements encodes the transposase that is required for transposition. The difference between the left element of Tn5, which is called IS50-L, and the right element, IS50-R, is a single nucleotide change. This creates an ochre mutation in the genes both for the transposase and for a second protein that is translated in the same reading frame, and at the same time, creates a promoter pointed to the right.
Transposition by members of the Tn5-Tn10 class of
transposons appears to require at least one protein encoded by the transposon
and only DNA sites located very near the ends of the element. If internal
sequences are deleted and if the necessary proteins are provided in trans, the deleted element will
transpose, although at a very low rate, for the transposase from Tn10 is
exceedingly unstable. This protein functions far better in cis, and even has trouble translocating longer versions of the
transposon because of the distance separating its ends. Transposons in this
class cut themselves out of one location in the chromosome and insert
themselves into another. Their main problem then becomes one of arranging that
such a repositioning ultimately results in a net gain in the number of
transposon copies.
Tn3 belongs to a second class of transposons.
Members of this group do not contain flanking IS sequences and possess only
short inverted repeats at their ends. Their transposition requires two
transposon-en-coded proteins—a transposase and another protein. Moreover,
transpo-sition by these elements requires more than the two proteins and the
ends of the transposon. A DNA site in the middle of the transposon is
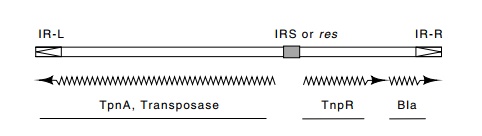
also required. Without this site, the transposon
does not complete transposition events; it copies itself into new locations but
connects the old location to the new (Fig. 19.7). Consequently, a transposon
lacking the internal site fuses the donor and target DNA replicons when it
attempts transposition. These properties demonstrate that transposi-tion by
these transposons occurs by copying rather than by excision and reintegration.
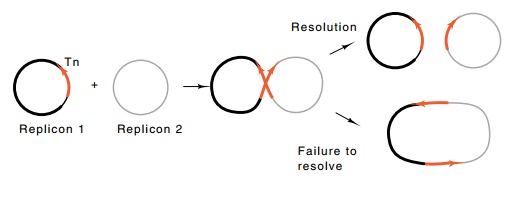
Figure
19.7 Incomplete transposition can fuse
two replicons with the dupli-cation of the transposition.
The third class of transposon is represented by
phage Mu. Superfi-cially this can be thought of as a transposon that happens to
have picked up a phage. Therefore it replicates not by freeing itself from the
chro-mosome but by copying itself into multiple locations. This property is
particularly useful for study of the process because instead of transpos-ing at
a frequency of perhaps 10- 4 per generation like many transposons, when
induced, it transposes at a frequency of 102 per cell doubling time.
Tn7 constitutes a fourth class of transposon. It
transposes into a unique site on the E.
coli chromosome with high specificity, but if this site is absent, the
transposon integrates nearly at random throughout the chromosome.
Related Topics