Chapter: Genetics and Molecular Biology: Transposable Genetic Elements
Inverting DNA Segments by Recombination, Flagellin Synthesis
Inverting DNA Segments by Recombination,
Flagellin Synthesis
Inversion of a stretch of DNA and excision of a
stretch of DNA, which will be discussed later, are closely related. Both
involve crossover events at the boundaries of the segment. As we saw, (Fig.
18.19), site-specific recombination between two recognition re-gions on a DNA
molecule can invert the intervening DNA. If one of the recognition regions is
reversed, however, the same excision reaction will excise the DNA.
Here we will be concerned with a form of regulation
that involves inverting a DNA segment. Later, a full chapter will be devoted to
chemotaxis, the ability of cells to sense and move toward attractants or away
from repellents. The movement is generated by rotation of flagella. In Salmonella two different genes encode
flagellin. Depending on the
orientation of an invertible controlling DNA
element, only one or the other of the flagellin genes is expressed at one time.
In the habitat of Salmonella, the gut, considerable selective advantage accrues to
the strain that can express more than one flagellin type. Antibody secreted by
patches of special cells on the walls of the intestine can bind to a flagellum
and block rotation of the flagella and therefore stop chemotaxis. Hence, to
survive and swim in the presence of antiflag-ellin antibody, it is to a cell’s
advantage to be able to synthesize a second type of flagellin against which the
host does not yet possess antibody. In these circumstances, the simultaneous
synthesis of two types of flagellin within a cell would be harmful. If both
types of flagellin were incorporated into flagella, then either of two types of
antibody could block chemotaxis rather than just one. Consequently, if two
flagellin genes are present, one should be completely inactive while the other
is functioning.
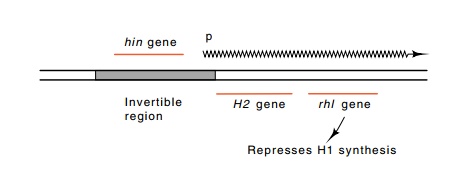
Figure
19.8 The structure of the DNA at the
flagellin controlling sequence inSalmonella.
The hin gene codes for a protein that
can invert the controllingregion. The region also contains a promoter for
synthesis of H2 and rhl messenger when the region is in one
orientation.
How can an appropriate flagellin synthesis
flip-flop circuit be con-structed using regulatory elements available to a
cell? Only one of the flagellin genes should possess any activity at one time,
but reversing the activity pattern should be readily possible. These requirements
are not easily met using regulatory elements of the type we have thus far
considered. In general, the regulation systems we have considered in previous
chapters do not permit construction of simple circuits possess-ing two states
of dramatically different activities and at the same time permit simple
switching on time scales of hours between the two states. For example, although
the lambda phage early gene system possesses the necessary selectivity in
having two states, fully off and fully on, this system is not reversible.
Alternatively, genes regulated like lac,
trp, and ara either do not possess large differences between their on and
offstates, or they flip states on time scales short compared to a doubling time
of the cells.
An invertible controlling segment of DNA solves Salmonella’s prob-lem. In one
orientation, only one type of flagellin is synthesized, and in the other
orientation, only the other type is synthesized. The apparatus responsible for
flipping the DNA segment can be completely inde-pendent of flagellin or
expression of the flagellin genes. That is, the protein or proteins necessary
for inversion of the DNA segment can be synthesized independently at a rate
appropriate for optimum survival of the population.
The structure of the invertible segment hin in Salmonella is similar to IS elements and also to the portion of Mu phage called the G segment. The hin segment is 995 base pairs long. It has inverted repeats of 14 bases at its ends, and its central region encodes a protein that is necessary for inversion. At its right end the element contains a promoter oriented to the right. Just to the right of the invertible segment lies one of the flagellin genes, H2, and another gene called rhl. The Rhl protein is a repressor of the second flagellin gene, H1, located elsewhere on the chromosome. As a result of this structure, one orientation of the invert-ible element provides synthesis of H2 protein and Rhl repressor. In this state the Rhl repressor blocks synthesis of H1 flagellin (Fig. 19.8). When the controlling element is in the other orientation, H2 and Rhl protein are not synthesized because they have been separated from their pro-moter, and therefore synthesis of H1 is not repressed.
The strategy of switching the antigenic properties
of crucial surface structures is not unique to bacteria. African trypanosomes,
unicellular flagellated cells, can evade the immune systems of vertebrates by
switch-ing their synthesis of surface proteins. These protozoa have hundreds of
different genes for the same basic surface protein. The process of switching on
the synthesis of any one of them, however, resembles more closely the
mating-type switch of yeast than the orientation switching involved with the
switching of Salmonella flagellin
synthesis.
Related Topics