Chapter: Genetics and Molecular Biology: Transposable Genetic Elements
Retrotransposons in Higher Cells
Retrotransposons in Higher Cells
The genomes of higher cells contain substantial
numbers of repeated sequences. For example, the Alu sequence of 300 base pairs constitutes about 5% of the human
genome. Among these repeated sequences are two main classes of transposable
elements. One is similar to Tn10 in its DNA transposition mode. This includes
the Ac element in maize, Tc1 in nematodes, and the P element in Drosophila. Members in the other class
transpose by means of an RNA intermediate, and they include the Ty1 factor in
yeast, the copia-like elements in Drosphila
and the long inter-spersed, LI elements in mammals. These elements are
retrotransposons and are closely related to retroviruses, if not identical to
retroviruses in some cases.
Hybridization and sequencing of Ty1 and the regions
into which it inserts have revealed its structure. It duplicates five bases
upon insertion
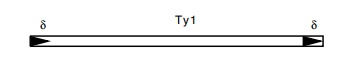
and consists of two flanking regions, called delta,
of 330 base pairs oriented as direct repeats around a 5,600-base-pair central
region that contains considerable homology to retroviruses. Not all delta
elements found in yeast are identical, nor are the central regions identical,
for some Ty elements are able to block expression of nearby yeast genes while
others stimulate expression of genes near the point of integration.
A recombination event between the two delta
elements deletes the central region and one delta element. Hence it is not
surprising that the yeast genome contains about 100 of these solo delta
elements. Recombination between different Ty elements can create various
chromosomal rearrangements. Although the consequences of recombination can be
determined easily in yeast, similar chromosome rearrangements catalyzed by
recombination between repeated sequences must also occur in other organisms.
Consequently transposons may be of positive value to an organism because they
may speed chromosome rearrangements that may directly generate new proteins
and new schemes of gene regulation.
Retroviruses, whose study began long before their
discovery as a part of the yeast Ty1 factors, are particles that contain
single-stranded RNA. This is both translated upon infection as well as
converted via an RNA-DNA hybrid to a DNA-DNA duplex that is often integrated
into the genome of the infected cell and is called a provirus. This form of the
virus and defective variants can be considered the retrotransposon. The
generation of the RNA-DNA duplex is catalyzed by reverse transcriptase. This
enzyme is packaged within the virus particle. Upon their insertion into the
chromosome, retroviruses duplicate a small number of bases as a result of
generating staggered nicks in the target sequence. At each end of the
retrovirus sequence is an inverted repeat of about 10 base pairs that is part
of a few-hundred-base-pair direct repeat. Between the direct repeats, which are
named long terminal repeats or LTRs, are sequences of about 5,000 base pairs
that code for viral coat protein and other proteins. Transcription begins near
the end of one LTR, proceeds
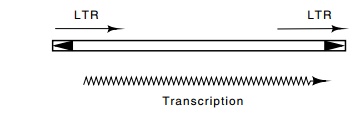
Some of the DNA that is found in repeated sequences
in Drosophila has been analyzed, and
its properties suggest that it too is related to proviruses. One such family is
copia. Small virus-like particles
contain-ing RNA homologous to copia
DNA sequences can even be isolated from the nucleus of Drosophila cells. This RNA is translatable into one of the proteins
that coats the RNA.
Retroviruses have been much harder to demonstrate in humans than in other animals. Nonetheless, they have been found. One was found to have inserted a copy of itself into the gene encoding factor VIII which is necessary for blood coagulation.
A more easily observed element in the human genome
is the Alu family of sequences.
Humans contain 100,000 to 500,000 copies of this sequence. The name derives
from the fact that the restriction enzyme Alu
cleaves more than once within the sequence. Consequently, diges-tion of
human DNA with Alu yields 100,000
identical fragments, which upon electrophoresis generate a unique band in
addition to the faint smear generated by the heterogeneity of the remainder of
the DNA. The Alu sequences look like
direct DNA copies of mRNA molecules becausethey contain a stretch of poly deoxyadenosine
at their 3’ ends. Like transposons, the Alu
sequences also are flanked by direct repeats of chromosomal sequences of 7 to
20 base pairs. It is not possible to tell from their structure whether
retroviruses and Alu sequences
evolved from transposable elements or the reverse.
Related Topics