Chapter: Genetics and Molecular Biology: Transposable Genetic Elements
Discovery of Tn Elements
Discovery of Tn Elements
While IS elements were under investigation, another
category of genetic elements was also being studied. These were plasmids that
carry genes encoding proteins that confer resistance to antibiotics. They are
called R-factors.
Not long after antibiotics began to be widely used,
many bacterial isolates from human infections were found to be resistant to one
or more of the drugs. These isolates were not resistant by virtue of a mutation
altering the cellular target of the antibiotics; instead, they synthesized
specific proteins or enzymes, which conferred resistance to the cells either by
detoxifying the antibiotics or by blocking their entry into the cell. For
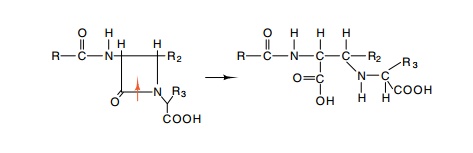
example, penicillin-resistant strains were found to synthesize a β-lactamase that opens the lactam ring of penicillin
and renders it harmless to the cells (Fig. 19.5).
Although many R-factors were fertile and could
transfer themselves to other cells by conjugation, the speed with which
different drug resistances appeared on R-factors was astonishingly rapid. Soon
after the introduction of a new antibiotic, R-factors that carried genes
con-ferring resistance to the new antibiotic would be found in many geographic
locations. Unfortunately this has created great problems in the treatment of
infections. The drug-resistance factors rapidly pick up genes encoding
resistance to the most commonly used drugs, and soon treatment of infections
becomes difficult. Now physicians use restraint in prescribing antibiotics to
slow the spread of drug-resistance genes.
The ability of drug-resistance genes to spread
rapidly suggested that they could hop from one R-factor to another or to other
DNA sequences such as phage or the bacterial chromosome. One demonstration of
this property came from an attempt to construct a lambda phage carrying the
kanamycin-resistance gene from an R-factor.
The objective of the experiment was to generate a λkan transducing phage by forcing lambda to insert at
random locations, and then excise. A small fraction of the excised phage would
have picked up the kanamy-cin genes and be able to transduce cells to Kanr. Cells
deleted of attB and carrying a
kanamycin-resistance R-factor were infected with lambda. This lambda phage was
deleted of part of the b2 region so
that the DNA of the resulting kanamycin-transducing phage would not be
Figure
19.5 The structure of the lactam ring
of penicillins and related antibi-otics and their structure following ring
opening by β-lactamase. Different penicillins
possess different R groups.
too large to be packaged. To permit the phage to
integrate via the Int pathway, the deletions chosen did not extend into the att region. As seen, a small fraction of
the resulting lysogens should then contain a lambda inserted into DNA adjacent
to the kanamycin-re-sistance gene(s) by virtue of integrating into sites that
weakly resemble lambda’s normal integration site (Fig. 19.6). Upon induction, a
small fraction of the phage were expected to excise incorrectly, pick up the
adjacent kanamycin-resistance genes, and be capable of transducing kanamycin
resistance to other cells.
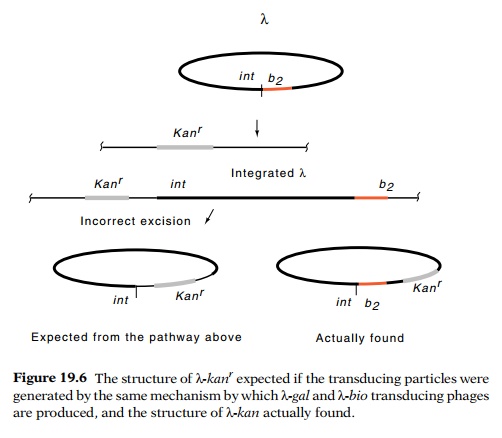
Indeed, kanamycin-transducing phage were isolated.
The phage pos-sessed several unexpected properties, however. First, the
additional DNA in these phage was not located immediately adjacent to attP. Second, kanamycin-resistant
transformants obtained by infecting cells with the λkan phage were lysogenic for lambda only if they
contained an attB. These properties
can be understood as follows. The DNA coding for kanamycin resistance
originally hopped or was copied onto lambda by a recombination event rather than
arriving there after an improper phage excision of the type that produces gal and bio transducing parti-cles. Therefore the foreign DNA could be
located anywhere on the phage and need not be immediately adjacent to attP. Second, a copy of the sequence encoding
the kanamycin resistance could also hop or be copied from the phage onto the
chromosome.
The DNA responsible for conferring the kanamycin resistance was examined by DNA heteroduplexes. It was found to be 5,400 base pairs long and flanked by inverted repeated sequences 1,500 base pairs long. Some of the drug-resistance genes studied on the R-factors themselves
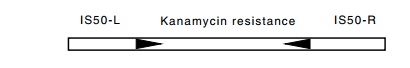
also possessed similar flanking sequences. Further
study showed that the repeated sequences flanking the drug-resistance genes
were trans-ferred when the drug-resistance gene hopped. Finally, it was found
that the repeated elements at the ends of many of the drug-resistance genes
possessed DNA sequences essentially identical to known IS elements. Thus it
appears that two IS elements can flank a gene and then facilitate copying the
composite element into other DNA locations by the same transposition reactions
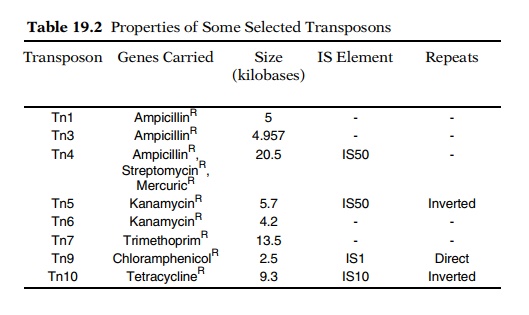
as are used by IS elements themselves. Se-quences with these general properties
and structure are called transposons, or Tn elements (Table 19.2). The element
described in this section is called Tn5, and the IS sequences associated with
it are called IS50 so as not to be confused with IS5.
Related Topics