Chapter: Genetics and Molecular Biology: Transposable Genetic Elements
Structure and Properties of IS Elements
Structure and Properties of IS Elements
Most of the bacterial IS sequences contain two
characteristic elements. First, a sequence of 10 to 40 base pairs at one end is
repeated or nearly repeated in an inverse orientation at the other end. This is
called IR, for inverted repeat. Second, the IS sequences contain one or two
regions that appear to code for protein because they possess promoters,
ribo-some-binding sequences, ATG (AUG) codons, and sense codons fol
If
proteins were made under control of these sequences, they would likely be
involved with the process of copying the IS into a new position in the process
of transpo-sition. Indeed, one of the gene products is generally called the
transposase. As only extremely low quantities of a transposase are needed, its
transcription and translation are specially designed to be inefficient.
Additionally, some premium is placed on possessing a mini-mum size, and genes
of a transposon frequently overlap. For example, in IS5 two oppositely oriented
reading frames overlap. The shorter is contained entirely within the longer
(Fig. 19.3).
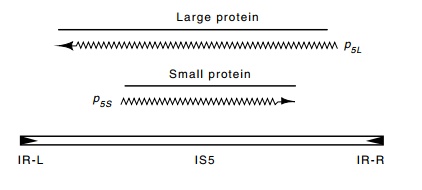
Figure 19.3 The organization of IS5. The large and small proteins are in thesame codon register.
A high expression of the IS genes most likely would
stimulate a high rate of transposition into new chromosomal sites. Since IS
transposition rates appear low, something must protect IS genes from
readthrough transcription and translation if the sequence has inserted into an
active gene of the host cell. IS5 possesses six peptide chain termination
codons flanking the protein genes—one at each end of the transposon, one in
each reading frame. Additionally, the left end of the IS5 contains a
transcription termination sequence oriented so as to prevent transcrip-tion
from adjacent bacterial genes from entering the IS in the reading direction of
the transposase.
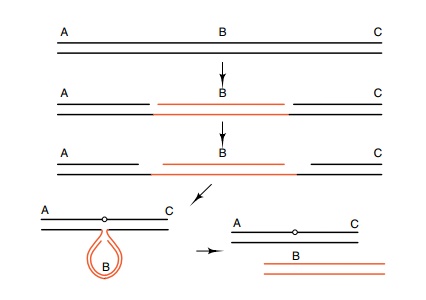
Figure 19.4 One mechanism by which nicking DNA can stimulate generationof deletions.
Not only do some IS sequences copy themselves or
jump into new DNA locations, but they also stimulate the creation of deletions
located near the IS elements. These deletions have one end precisely located at
the end of an IS element and apparently arise in abortive transposition attempts
that begin with transposase nicking the DNA at one of the boundaries of the IS
element (Fig. 19.4). Such a facile creation of deletions in any selected region
is highly useful to geneticists, for, as we have seen, a set of such deletions
can be used for fine structure mapping.
Related Topics