Chapter: Basic Concept of Biotechnology : Tools and Techniques in Biotechnology
Southern Blotting Technique - Techniques of Biotechnology and Innovations
SOUTHERN
BLOTTING TECHNIQUE
The
technique is useful in confirming the cloned fragment in the plasmid/ genomic
DNA of corresponding organism. The template genomic DNA molecule fragment with
a defined size by restriction enzymes and immobilized on nitrocellulose
membrane. The labeled probe is allowed to hybridize with complementary fragment
which is immobilized on the membrane. The technique includes 4-5 steps viz.,
isolation of DNA, blotting technique, DNA/DNA hybridization, washing and
visualization (Fig. 3).
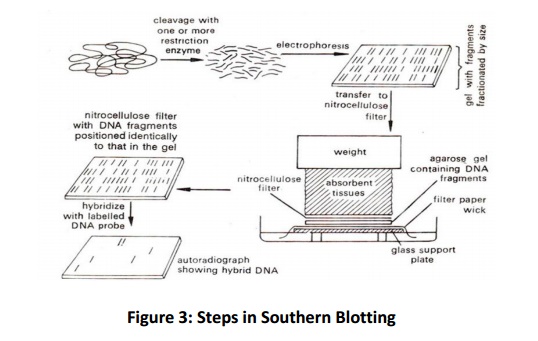
The
DNA is treated with one or more restriction enzymes and fragments are separated
on the agarose gel. Before transferring to nylon/nitrocellulose membrane the
double strands fragmented DNA is denatured by alkaline treatment in order to
facilitate the immobilization and proper hybridization with the probe. If the
nitrocellulose membrane is being used for blotting than after alkaline
treatment soak the gel in Tris salt buffer, because under highalkaline
condition (pH 9.0) the DNA fragments will not bind to nitrocellulose membrane.
The prepared gel placed on top of the buffer saturated filter paper, then thin
layer of nitrocellulose paper on top of the gel and finally some dry filter
paper kept over the nitrocellulose membrane. Some wait will be kept on the top
to facilitate good capillary action. The entire assembly kept in buffered
solution but only lower filter paper should touches to the buffer, it will
build the capillary action and from the top dry paper receive the buffer.
During capillary movement, the DNA transferred from gel to membrane, in the
same fashion as they are present in the gel. This transfer buffer varies with
type of membrane used, for nitrocellulose membrane the high-salt transfer
buffer (20 X SSC, which comprises 3.0 mol/L NaCl and 0.3 mol/L sodium citrate)
and for nylon membrane alkaline transfer buffer (0.4 mol/L NaOH) is used. The
transfer process takes for 18 h in case of SSC buffer and 2 h in alkaline transfer
buffer. After the transfer process gently rinse the membrane in 2X SSC buffer
and air dry. At this step the DNA is loosely bound to nitrocellulose membrane
hence, it is permanently immobilized by baking the membrane at 80oC
for 2 hr. The free space in membrane is blocked with 0.2% each of Ficoll,
polyvinyl pyrrolidone and bovine serum albumin, the process is called as
prehybridization, runs for 15 min to 3 h at 65oC. Alternatively,
this mixture can be included in the hybridization buffer. Later treat the
membrane with hybridization buffer 2 X SSC containing 1% SDS and labelled
(radioactive labelled/ fluorescent labelled) probe.
Keep
it in agitating condition or in a tube that is constantly rotated so all parts
of the membrane are exposed to the probe. During the hybridization chances of
non-specific annealing, it can be reduced by using the buffer of high ionic
concentration and incubating the hybridization process at/just below melting
temperature of probe and template. After hybridization, the membrane is washed
and subjected for detection procedure either by X-ray in case of radioactive
labelled probe or naked visualization for fluorescent labelled probe.
Related Topics