Chapter: Genetics and Molecular Biology: Transcription,Termination, and RNA Processing
Polymerase Elongation Rate
Polymerase Elongation Rate
Even more than in DNA synthesis, it is sensible for
cells to regulate RNA synthesis at the initiation steps so that the elaborate
machinery involved in independently regulating thousands of genes need not be
built into the basic RNA synthesis module. Once RNA synthesis has been
initiated, it proceeds at the same average rate on most, independent of growth
conditions. Can this be demonstrated? Another need for knowing the RNA
elongation rate is in the interpretation of physiological experiments. How
soon after the addition of an inducer can a newly synthe-sized mRNA molecule
appear?
RNA elongation rate measurements are not too hard to perform invitro, but they are appreciably more difficult to perform on growing cells. Here we shall explain one method that has been used to determine the in vivo RNA elongation rate in Escherichia coli.
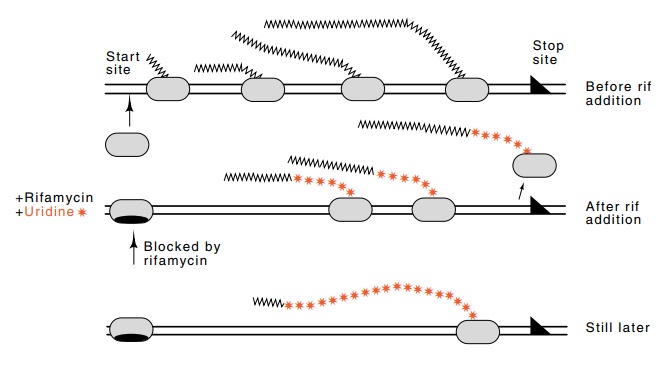
Figure
5.1 Effects of rifamycin addition on
transcription of a large operon.Upon the addition of rifamycin, no more RNA
polymerase molecules may initiate transcription. Those polymerase molecules
that were transcribing con-tinue to the end of the operon. Finally, the
polymerase molecule that had initiated transcription just before the addition
of rifamycin completes tran-scription of the operon.
The measurement used rifamycin, an antibiotic that
inhibits RNA polymerase only at the initiation step. It has no effect on RNA
polym-erase molecules engaged in elongation. Rifamycin and radioactive urid-ine
were simultaneously added to bacteria; thus only those RNA chains that were in
the process of elongation at the time of the additions were radioactively
labeled, and no new ones could be initiated (Fig. 5.1). At various times after
the rifamycin and uridine addition, samples were taken from the culture and
their RNA was separated according to size
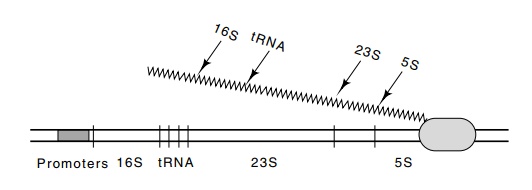
Figure
5.2 Structure of the ribosomal RNA
operon used to determine the RNAelongation rate in E. coli.
by electrophoresis on polyacrylamide gels. Suppose
that a specific species of RNA molecule is well separated from all other
species by the electrophoresis. Then, the radioactivity in this size class will
increase with time for as long as RNA polymerase molecules transcribe the
corresponding gene, but once the last polymerase molecule to initiate has
crossed the region, there can be no additional increase in radioac-tivity. The
interval between the addition of rifamycin and the end of the period over which
radioactivity increases is the time required for an RNA polymerase molecule to
transcribe from the promoter to the end of the transcribed region.
The ribosomal RNA gene complexes were a convenient
system for these measurements. Each of these seven nearly identical gene
com-plexes consists of two closely spaced promoters, a gene for the 16S
ribosomal RNA, a spacer region, a tRNA gene, the gene for the 23S ribosomal
RNA, and the gene for the 5S ribosomal RNA (Fig. 5.2). The total length of this
transcriptional unit is about 5,000 nucleotides. The 16S RNA, spacer tRNA, 23S
RNA, and 5S RNA are all generated by cleavage from the growing polynucleotide
chain.
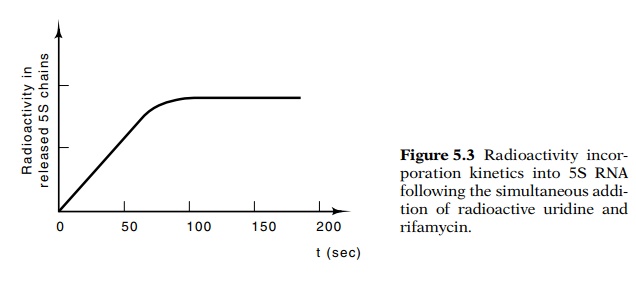
The interval between the time of rifamycin addition
and the time at which the last RNA polymerase molecule transcribes across the
end of the 5S gene is the time required for RNA polymerase to transcribe the
5,000 bases from the promoter to the end of the ribosomal gene com-plex. This
time is found from the radioactive uridine incorporation measurements.
Transcription across the 5S gene ends when the radio - activity in 5S RNA stops
increasing. This happens about 90 seconds after rifamycin and uridine addition
(Fig. 5.3). This yields an elongation rate of about 60 nucleotides per second.
This type of elongation rate meas-urement has been performed on cells growing
at many different growth rates, and as expected, the results show that the RNA
chain growth rate is independent of the growth rate of cells at a given
temperature.
Related Topics