Chapter: Genetics and Molecular Biology: Transcription,Termination, and RNA Processing
Caps, Splices, Edits, and Poly-A Tails on Eukaryotic RNAs
Caps, Splices, Edits, and Poly-A Tails on
Eukaryotic RNAs
Eukaryotic RNAs are processed at their beginning,
middle, and terminal sections. Although the structures resulting from these
posttranscrip-tional modifications are known, why the modifications occur is
not clear.
Figure 5.11 The structure of the cap found on eukaryotic messenger RNAs.The first base is 7-methylguanylate connected by a 5’-5’ triphosphate linkage to the next base. The 2’ positions on bases 1 and 2 may or may not be methylated.
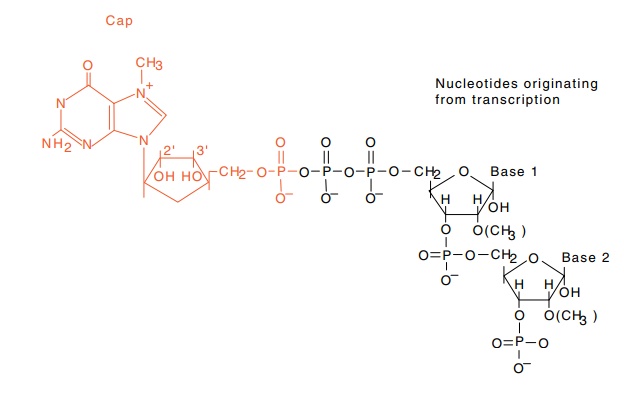
The 5’ ends of nearly all cellular messenger RNAs
contain a “cap,” a guanine in a reversed orientation plus several other
modifications (Fig. 5.11). More precisely, the cap is a guanine methylated on
its 7 position joined through 5’-5’ pyrophosphate linkage to a base derived
from transcription. Both the first and second bases after the capping
nucleo-tide usually are methylated. These modifications were discovered by
tracking down why and where viral RNA synthesized in vitro could be methylated. The trail led to the 5’ end whose
structure was then deter-mined by Shatkin. The cap helps stabilize the RNA, is
sometimes involved in export to the cytoplasm, may be involved with splicing,
and also assists translation. Ribosomes translate cap-containing messengers
more efficiently than messengers lacking a cap.
A sizable fraction of the RNA that is synthesized
in the nucleus of eukaryotic cells is not transported to the cytoplasm. This
RNA is of a variety of sizes and sequences and is called heterogeneous nuclear
RNA. Within this class of RNA are messenger RNA sequences that ultimately are
translated into protein, but, as synthesized, the nuclear RNAs are not directly
translatable (Fig. 5.12). They contain extraneous sequences that must be
removed by cutting and splicing to form the continuous
Figure
5.12 Maturation of aprimary transcript
to yield a translatable message.
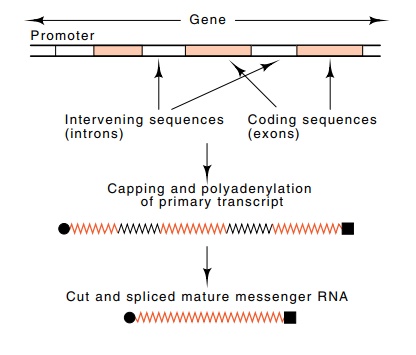
coding sequence that is found in the mature
messenger in the cytoplasm. The intervening sequences, which are removed from
mature RNA, are called introns, and the other parts of the messenger sequence
are often referred to as exons.
Another type of modification in RNA after its
synthesis is called editing. Although most RNAs are spliced, editing occurs
very rarely. One prominent example of editing is the apolipoprotein. Humans
contain a single copy of this gene. In the liver its gene product is 512,000
daltons molecular weight, but in intestinal cells its gene product is only
242,000 daltons molecular weight. Examination of the mRNA shows that a specific
cytosine of the RNA is converted to a uracil or uracil-like nucleotide in
intestinal cells, but not in hepatic cells. This conversion creates a
translation stop codon, hence the shorter gene product in intestinal cells. The
conversion process involved is called RNA editing. It is also found in some
animal viruses. In a few extreme cases, hundreds of nucleotides in an RNA are
edited. In these cases, special short guide RNA molecules direct the choice of
inserted or deleted nucleotides.
The final type of RNA modification found in
eukaryotic cells is the posttranscriptional addition to the 3’ end of 30 to 500
nucleotides of polyadenylic acid. This begins about 15 nucleotides beyond the
poly-A signal sequence AAUAAA. Transcription itself appears to terminate
somewhat beyond the poly-A signal and processing quickly removes the extra
nucleotide before the poly-A addition.
Related Topics